Systemic Expression, Purification, and Initial Structural Characterization of Bacteriophage T4 Proteins Without Known Structure Homologs
- PMID: 33927712
- PMCID: PMC8076793
- DOI: 10.3389/fmicb.2021.674415
Systemic Expression, Purification, and Initial Structural Characterization of Bacteriophage T4 Proteins Without Known Structure Homologs
Abstract
Bacteriophage T4 of Escherichia coli is one of the most studied phages. Research into it has led to numerous contributions to phage biology and biochemistry. Coding about 300 gene products, this double-stranded DNA virus is the best-understood model in phage study and modern genomics and proteomics. Ranging from viral RNA polymerase, commonly found in phages, to thymidylate synthase, whose mRNA requires eukaryotic-like self-splicing, its gene products provide a pool of fine examples for phage research. However, there are still up to 130 gene products that remain poorly characterized despite being one of the most-studied model phages. With the recent advancement of cryo-electron microscopy, we have a glimpse of the virion and the structural proteins that present in the final assembly. Unfortunately, proteins participating in other stages of phage development are absent. Here, we report our systemic analysis on 22 of these structurally uncharacterized proteins, of which none has a known homologous structure due to the low sequence homology to published structures and does not belong to the category of viral structural protein. Using NMR spectroscopy and cryo-EM, we provided a set of preliminary structural information for some of these proteins including NMR backbone assignment for Cef. Our findings pave the way for structural determination for the phage proteins, whose sequences are mainly conserved among phages. While this work provides the foundation for structural determinations of proteins like Gp57B, Cef, Y04L, and Mrh, other in vitro studies would also benefit from the high yield expression of these proteins.
Keywords: Cef; Mrh; NMR; T4; Y04L and Gp57B; bacteriophage; cryo-EM.
Copyright © 2021 Zhang, Li, Wang, Li, Ma, Chen, Li, Yang, Wang and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
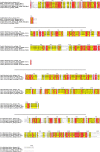


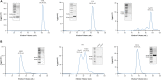

Similar articles
-
Resonance assignments of bacteriophage T4 Y04L protein.Biomol NMR Assign. 2020 Apr;14(1):51-54. doi: 10.1007/s12104-019-09919-5. Epub 2019 Nov 9. Biomol NMR Assign. 2020. PMID: 31707562
-
Sinorhizobium meliloti Phage ΦM9 Defines a New Group of T4 Superfamily Phages with Unusual Genomic Features but a Common T=16 Capsid.J Virol. 2015 Nov;89(21):10945-58. doi: 10.1128/JVI.01353-15. Epub 2015 Aug 26. J Virol. 2015. PMID: 26311868 Free PMC article.
-
The genome of bacteriophage phiKZ of Pseudomonas aeruginosa.J Mol Biol. 2002 Mar 15;317(1):1-19. doi: 10.1006/jmbi.2001.5396. J Mol Biol. 2002. PMID: 11916376
-
Baseplate assembly of phage Mu: Defining the conserved core components of contractile-tailed phages and related bacterial systems.Proc Natl Acad Sci U S A. 2016 Sep 6;113(36):10174-9. doi: 10.1073/pnas.1607966113. Epub 2016 Aug 23. Proc Natl Acad Sci U S A. 2016. PMID: 27555589 Free PMC article.
-
Bacteriophage-encoded functions engaged in initiation of homologous recombination events.Crit Rev Microbiol. 2009;35(3):197-220. doi: 10.1080/10408410902983129. Crit Rev Microbiol. 2009. PMID: 19563302 Review.
Cited by
-
Circular single-stranded DNA as a programmable vector for gene regulation in cell-free protein expression systems.Nat Commun. 2024 May 31;15(1):4635. doi: 10.1038/s41467-024-49021-6. Nat Commun. 2024. PMID: 38821953 Free PMC article.
-
Integrated Omics Reveal Time-Resolved Insights into T4 Phage Infection of E. coli on Proteome and Transcriptome Levels.Viruses. 2022 Nov 12;14(11):2502. doi: 10.3390/v14112502. Viruses. 2022. PMID: 36423111 Free PMC article.
-
Bacteriophage protein Gp46 is a cross-species inhibitor of nucleoid-associated HU proteins.Proc Natl Acad Sci U S A. 2022 Mar 1;119(9):e2116278119. doi: 10.1073/pnas.2116278119. Proc Natl Acad Sci U S A. 2022. PMID: 35193978 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

