Risk-Based Chemical Ranking and Generating a Prioritized Human Exposome Database
- PMID: 33929905
- PMCID: PMC8086799
- DOI: 10.1289/EHP7722
Risk-Based Chemical Ranking and Generating a Prioritized Human Exposome Database
Abstract
Background: Due to the ubiquitous use of chemicals in modern society, humans are increasingly exposed to thousands of chemicals that contribute to a major portion of the human exposome. Should a comprehensive and risk-based human exposome database be created, it would be conducive to the rapid progress of human exposomics research. In addition, once a xenobiotic is biotransformed with distinct half-lives upon exposure, monitoring the parent compounds alone may not reflect the actual human exposure. To address these questions, a comprehensive and risk-prioritized human exposome database is needed.
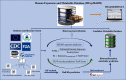
Objectives: Our objective was to set up a comprehensive risk-prioritized human exposome database including physicochemical properties as well as risk prediction and develop a graphical user interface (GUI) that has the ability to conduct searches for content associated with chemicals in our database.
Methods: We built a comprehensive risk-prioritized human exposome database by text mining and database fusion. Subsequently, chemicals were prioritized by integrating exposure level obtained from the Systematic Empirical Evaluation of Models with toxicity data predicted by the Toxicity Estimation Software Tool and the Toxicological Priority Index calculated from the ToxCast database. The biotransformation half-lives () of all the chemicals were assessed using the Iterative Fragment Selection approach and biotransformation products were predicted using the previously developed BioTransformer machine-learning method.
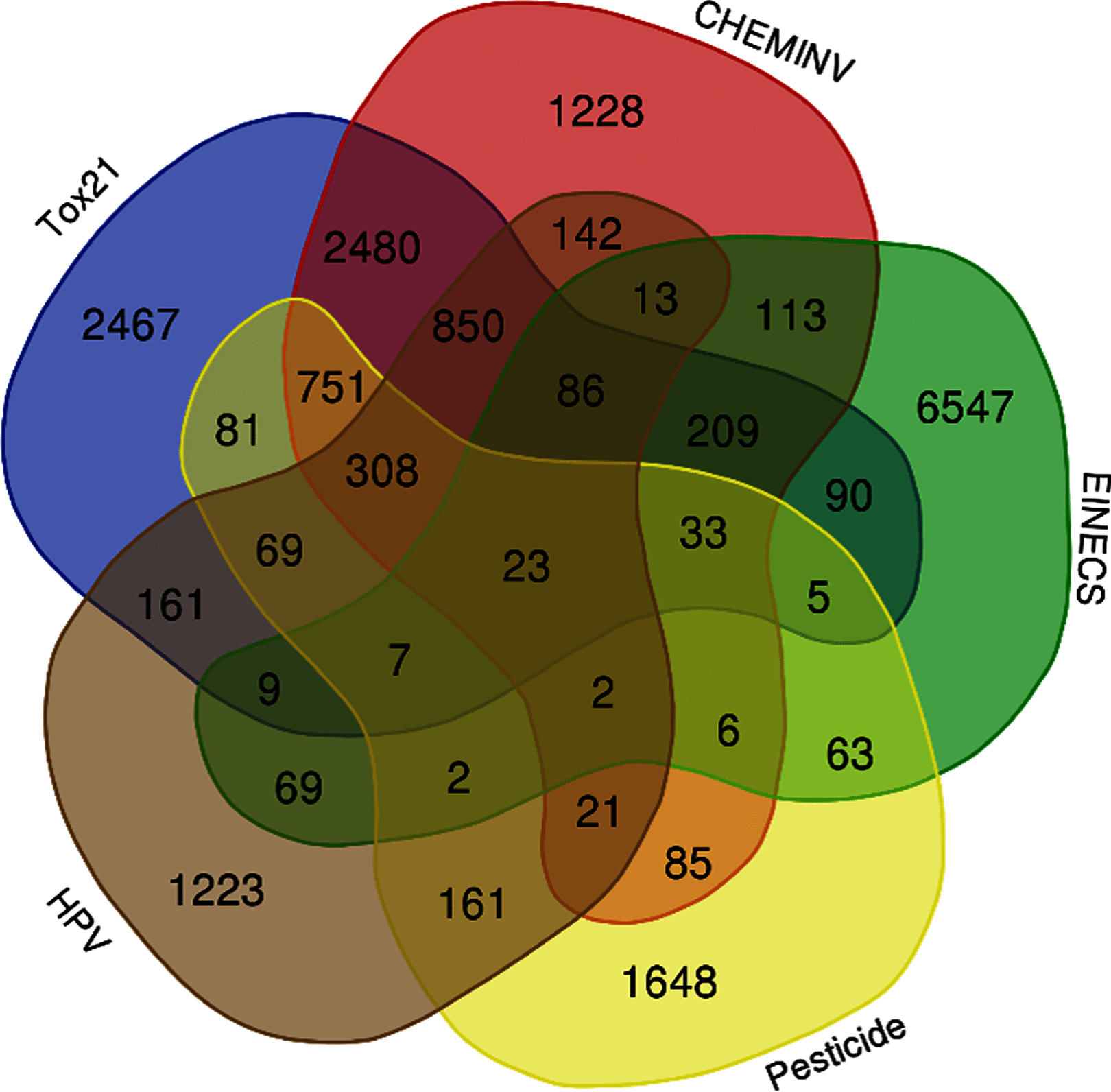
Results: We compiled a human exposome database of chemicals, prioritized 13,441 chemicals based on probabilistic hazard quotient and 7,770 chemicals based on risk index, and provided a predicted biotransformation metabolite database of metabolites. In addition, a user-interactive Java software (Oracle)-based search GUI was generated to enable open access to this new resource.
Discussion: Our database can be used to guide chemical management and enhance scientific understanding to rapidly and effectively prioritize chemicals for comprehensive biomonitoring in epidemiological investigations. https://doi.org/10.1289/EHP7722.
Figures
Similar articles
-
Generating the Blood Exposome Database Using a Comprehensive Text Mining and Database Fusion Approach.Environ Health Perspect. 2019 Sep;127(9):97008. doi: 10.1289/EHP4713. Epub 2019 Sep 26. Environ Health Perspect. 2019. PMID: 31557052 Free PMC article.
-
Human Indoor Exposome of Chemicals in Dust and Risk Prioritization Using EPA's ToxCast Database.Environ Sci Technol. 2019 Jun 18;53(12):7045-7054. doi: 10.1021/acs.est.9b00280. Epub 2019 May 28. Environ Sci Technol. 2019. PMID: 31081622 Review.
-
HExpPredict: In Vivo Exposure Prediction of Human Blood Exposome Using a Random Forest Model and Its Application in Chemical Risk Prioritization.Environ Health Perspect. 2023 Mar;131(3):37009. doi: 10.1289/EHP11305. Epub 2023 Mar 13. Environ Health Perspect. 2023. PMID: 36913238 Free PMC article.
-
An annotation database for chemicals of emerging concern in exposome research.Environ Int. 2021 Jul;152:106511. doi: 10.1016/j.envint.2021.106511. Epub 2021 Mar 24. Environ Int. 2021. PMID: 33773387
-
Toxicological and Exposure Database Inventory: A review.Int J Hyg Environ Health. 2022 Sep;246:114055. doi: 10.1016/j.ijheh.2022.114055. Epub 2022 Oct 21. Int J Hyg Environ Health. 2022. PMID: 36279787 Review.
Cited by
-
AirPen: A Wearable Monitor for Characterizing Exposures to Particulate Matter and Volatile Organic Compounds.Environ Sci Technol. 2023 Jul 25;57(29):10604-10614. doi: 10.1021/acs.est.3c02238. Epub 2023 Jul 14. Environ Sci Technol. 2023. PMID: 37450410 Free PMC article.
-
Reducing uncertainty in dose-response assessments by incorporating Bayesian benchmark dose modeling and in vitro data on population variability.Risk Anal. 2025 Feb;45(2):457-472. doi: 10.1111/risa.17451. Epub 2024 Aug 16. Risk Anal. 2025. PMID: 39148436 Free PMC article.
-
Investigating the origins of the mutational signatures in cancer.Nucleic Acids Res. 2025 Jan 7;53(1):gkae1303. doi: 10.1093/nar/gkae1303. Nucleic Acids Res. 2025. PMID: 39778866 Free PMC article. Review.
-
Probabilistic risk assessment - the keystone for the future of toxicology.ALTEX. 2022;39(1):3-29. doi: 10.14573/altex.2201081. ALTEX. 2022. PMID: 35034131 Free PMC article. Review.
-
In-utero exposure to real-life environmental chemicals disrupts gene expression within the hypothalamo-pituitary-gonadal axis of prepubertal and adult rams.Environ Res. 2025 Jan 1;264(Pt 1):120303. doi: 10.1016/j.envres.2024.120303. Epub 2024 Nov 5. Environ Res. 2025. PMID: 39510237 Free PMC article.
References
-
- Andrianou XD, van der Lek C, Charisiadis P, Ioannou S, Fotopoulou KN, Papapanagiotou Z, et al. . 2019. Application of the urban exposome framework using drinking water and quality of life indicators: a proof-of-concept study in Limassol, Cyprus. PeerJ 7:e6851, PMID: 31179170, 10.7717/peerj.6851. - DOI - PMC - PubMed
-
- Bland J. 2007. Managing biotransformation: introduction and overview. Altern Ther Health Med 13(2):S85–S87, PMID: 17405682. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

