A monoclonal antibody against staphylococcal enterotoxin B superantigen inhibits SARS-CoV-2 entry in vitro
- PMID: 33930306
- PMCID: PMC8082696
- DOI: 10.1016/j.str.2021.04.005
A monoclonal antibody against staphylococcal enterotoxin B superantigen inhibits SARS-CoV-2 entry in vitro
Abstract
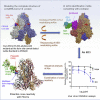
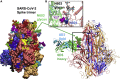
We recently discovered a superantigen-like motif sequentially and structurally similar to a staphylococcal enterotoxin B (SEB) segment, near the S1/S2 cleavage site of the SARS-CoV-2 spike protein, which might explain the multisystem inflammatory syndrome (MIS-C) observed in children and the cytokine storm in severe COVID-19 patients. We show here that an anti-SEB monoclonal antibody (mAb), 6D3, can bind this viral motif at its polybasic (PRRA) insert to inhibit infection in live virus assays. The overlap between the superantigenic site of the spike and its proteolytic cleavage site suggests that the mAb prevents viral entry by interfering with the proteolytic activity of cell proteases (furin and TMPRSS2). The high affinity of 6D3 for this site originates from a polyacidic segment at its heavy chain CDR2. The study points to the potential utility of 6D3 for possibly treating COVID-19, MIS-C, or common colds caused by human coronaviruses that also possess a furin-like cleavage site.
Keywords: 6D3; COVID-19; MIS-C; TMPRSS2; cytokine storm; furin-cleavage site; neutralizing antibodies; staphylococcal enterotoxin B; superantigen; viral entry.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Update of
-
A monoclonal antibody against staphylococcal enterotoxin B superantigen inhibits SARS-CoV-2 entry in vitro.bioRxiv [Preprint]. 2020 Nov 24:2020.11.24.395079. doi: 10.1101/2020.11.24.395079. bioRxiv. 2020. Update in: Structure. 2021 Sep 2;29(9):951-962.e3. doi: 10.1016/j.str.2021.04.005. PMID: 33269352 Free PMC article. Updated. Preprint.
Comment on
-
Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25254-25262. doi: 10.1073/pnas.2010722117. Epub 2020 Sep 28. Proc Natl Acad Sci U S A. 2020. PMID: 32989130 Free PMC article.
References
-
- Belhadjer Z., Méot M., Bajolle F., Khraiche D., Legendre A., Abakka S., Auriau J., Grimaud M., Oualha M., Beghetti M., et al. Acute heart failure in multisystem inflammatory syndrome in children (MIS-C) in the context of global SARS-CoV-2 pandemic. Circulation. 2020;142:429–436. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

