Functional landscape of SARS-CoV-2 cellular restriction
- PMID: 33930332
- PMCID: PMC8043580
- DOI: 10.1016/j.molcel.2021.04.008
Functional landscape of SARS-CoV-2 cellular restriction
Abstract
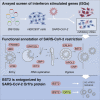
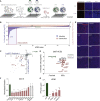
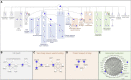
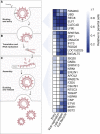
A deficient interferon (IFN) response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has been implicated as a determinant of severe coronavirus disease 2019 (COVID-19). To identify the molecular effectors that govern IFN control of SARS-CoV-2 infection, we conducted a large-scale gain-of-function analysis that evaluated the impact of human IFN-stimulated genes (ISGs) on viral replication. A limited subset of ISGs were found to control viral infection, including endosomal factors inhibiting viral entry, RNA binding proteins suppressing viral RNA synthesis, and a highly enriched cluster of endoplasmic reticulum (ER)/Golgi-resident ISGs inhibiting viral assembly/egress. These included broad-acting antiviral ISGs and eight ISGs that specifically inhibited SARS-CoV-2 and SARS-CoV-1 replication. Among the broad-acting ISGs was BST2/tetherin, which impeded viral release and is antagonized by SARS-CoV-2 Orf7a protein. Overall, these data illuminate a set of ISGs that underlie innate immune control of SARS-CoV-2/SARS-CoV-1 infection, which will facilitate the understanding of host determinants that impact disease severity and offer potential therapeutic strategies for COVID-19.
Keywords: BST2; ISG; Orf7a; SARS-CoV-2; innate immunity; interferon; viral evasion.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Update of
-
Functional Landscape of SARS-CoV-2 Cellular Restriction.bioRxiv [Preprint]. 2020 Sep 30:2020.09.29.319566. doi: 10.1101/2020.09.29.319566. bioRxiv. 2020. Update in: Mol Cell. 2021 Jun 17;81(12):2656-2668.e8. doi: 10.1016/j.molcel.2021.04.008. PMID: 33024967 Free PMC article. Updated. Preprint.
Comment in
-
Meet the authors: Laura Martin-Sancho and Sumit K. Chanda.Mol Cell. 2021 Jun 17;81(12):2497-2498. doi: 10.1016/j.molcel.2021.05.034. Mol Cell. 2021. PMID: 34143965 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- U19 AI168631/AI/NIAID NIH HHS/United States
- HHSN272201400008C/AI/NIAID NIH HHS/United States
- P41 GM103504/GM/NIGMS NIH HHS/United States
- U19 AI135990/AI/NIAID NIH HHS/United States
- R21 AI163912/AI/NIAID NIH HHS/United States
- P30 CA030199/CA/NCI NIH HHS/United States
- UL1 TR001422/TR/NCATS NIH HHS/United States
- P30 CA060553/CA/NCI NIH HHS/United States
- F31 AI141111/AI/NIAID NIH HHS/United States
- U19 AI135972/AI/NIAID NIH HHS/United States
- R37 AI081668/AI/NIAID NIH HHS/United States
- U19 AI118610/AI/NIAID NIH HHS/United States
- UL1 TR002389/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

