The AaCBF4-AaBAM3.1 module enhances freezing tolerance of kiwifruit (Actinidia arguta)
- PMID: 33931620
- PMCID: PMC8087828
- DOI: 10.1038/s41438-021-00530-1
The AaCBF4-AaBAM3.1 module enhances freezing tolerance of kiwifruit (Actinidia arguta)
Abstract
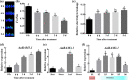
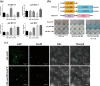
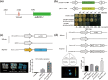
Beta-amylase (BAM) plays an important role in plant resistance to cold stress. However, the specific role of the BAM gene in freezing tolerance is poorly understood. In this study, we demonstrated that a cold-responsive gene module was involved in the freezing tolerance of kiwifruit. In this module, the expression of AaBAM3.1, which encodes a functional protein, was induced by cold stress. AaBAM3.1-overexpressing kiwifruit lines showed increased freezing tolerance, and the heterologous overexpression of AaBAM3.1 in Arabidopsis thaliana resulted in a similar phenotype. The results of promoter GUS activity and cis-element analyses predicted AaCBF4 to be an upstream transcription factor that could regulate AaBAM3.1 expression. Further investigation of protein-DNA interactions by using yeast one-hybrid, GUS coexpression, and dual luciferase reporter assays confirmed that AaCBF4 directly regulated AaBAM3.1 expression. In addition, the expression of both AaBAM3.1 and AaCBF4 in kiwifruit responded positively to cold stress. Hence, we conclude that the AaCBF-AaBAM module is involved in the positive regulation of the freezing tolerance of kiwifruit.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl Acad. Sci. USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. - DOI - PMC - PubMed
-
- Wisniewski M, Bassett C, Gusta LV. An overview of cold hardiness in Woody plants: Seeing the forest through the trees. Hortscience. 2003;38:952–959. doi: 10.21273/HORTSCI.38.5.952. - DOI
-
- Chen MJ, et al. Characterization of low temperature-induced plasma membrane lipidome remodeling combined with gene expression analysis reveals mechanisms that regulate membrane lipid desaturation in Carica papaya. Sci. Hortic. 2020;272:1–12.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

