Single nucleus RNA-sequencing defines unexpected diversity of cholinergic neuron types in the adult mouse spinal cord
- PMID: 33931636
- PMCID: PMC8087807
- DOI: 10.1038/s41467-021-22691-2
Single nucleus RNA-sequencing defines unexpected diversity of cholinergic neuron types in the adult mouse spinal cord
Abstract
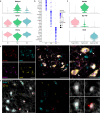
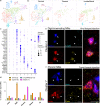
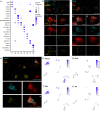
In vertebrates, motor control relies on cholinergic neurons in the spinal cord that have been extensively studied over the past hundred years, yet the full heterogeneity of these neurons and their different functional roles in the adult remain to be defined. Here, we develop a targeted single nuclear RNA sequencing approach and use it to identify an array of cholinergic interneurons, visceral and skeletal motor neurons. Our data expose markers for distinguishing these classes of cholinergic neurons and their rich diversity. Specifically, visceral motor neurons, which provide autonomic control, can be divided into more than a dozen transcriptomic classes with anatomically restricted localization along the spinal cord. The complexity of the skeletal motor neurons is also reflected in our analysis with alpha, gamma, and a third subtype, possibly corresponding to the elusive beta motor neurons, clearly distinguished. In combination, our data provide a comprehensive transcriptomic description of this important population of neurons that control many aspects of physiology and movement and encompass the cellular substrates for debilitating degenerative disorders.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

