MicroRNAs from extracellular vesicles as a signature for Parkinson's disease
- PMID: 33931970
- PMCID: PMC8021010
- DOI: 10.1002/ctm2.357
MicroRNAs from extracellular vesicles as a signature for Parkinson's disease
Conflict of interest statement
The authors declare no conflict of interest.
Figures
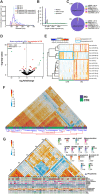
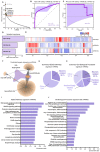
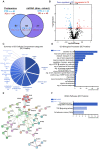
References
-
- Schlossmacher MG, Mollenhauer B. Biomarker research in Parkinson's disease: objective measures needed for patient stratification in future cause‐directed trials. Biomarkers Med. 2010;4:647‐650. - PubMed
-
- Saba R, Schratt GM. MicroRNAs in neuronal development, function and dysfunction. Brain Res. 2010;1338:3‐13. - PubMed
-
- Yagi Y, Ohkubo T, Kawaji H, et al. Next‐generation sequencing‐based small RNA profiling of cerebrospinal fluid exosomes. Neurosci Lett. 2017;636:48‐57. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
