Sub-genic intolerance, ClinVar, and the epilepsies: A whole-exome sequencing study of 29,165 individuals
- PMID: 33932343
- PMCID: PMC8206159
- DOI: 10.1016/j.ajhg.2021.04.009
Sub-genic intolerance, ClinVar, and the epilepsies: A whole-exome sequencing study of 29,165 individuals
Erratum in
-
Sub-genic intolerance, ClinVar, and the epilepsies: A whole-exome sequencing study of 29,165 individuals.Am J Hum Genet. 2021 Oct 7;108(10):2024. doi: 10.1016/j.ajhg.2021.08.008. Am J Hum Genet. 2021. PMID: 34626584 Free PMC article. No abstract available.
Abstract
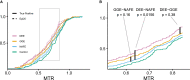
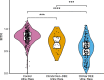
Both mild and severe epilepsies are influenced by variants in the same genes, yet an explanation for the resulting phenotypic variation is unknown. As part of the ongoing Epi25 Collaboration, we performed a whole-exome sequencing analysis of 13,487 epilepsy-affected individuals and 15,678 control individuals. While prior Epi25 studies focused on gene-based collapsing analyses, we asked how the pattern of variation within genes differs by epilepsy type. Specifically, we compared the genetic architectures of severe developmental and epileptic encephalopathies (DEEs) and two generally less severe epilepsies, genetic generalized epilepsy and non-acquired focal epilepsy (NAFE). Our gene-based rare variant collapsing analysis used geographic ancestry-based clustering that included broader ancestries than previously possible and revealed novel associations. Using the missense intolerance ratio (MTR), we found that variants in DEE-affected individuals are in significantly more intolerant genic sub-regions than those in NAFE-affected individuals. Only previously reported pathogenic variants absent in available genomic datasets showed a significant burden in epilepsy-affected individuals compared with control individuals, and the ultra-rare pathogenic variants associated with DEE were located in more intolerant genic sub-regions than variants associated with non-DEE epilepsies. MTR filtering improved the yield of ultra-rare pathogenic variants in affected individuals compared with control individuals. Finally, analysis of variants in genes without a disease association revealed a significant burden of loss-of-function variants in the genes most intolerant to such variation, indicating additional epilepsy-risk genes yet to be discovered. Taken together, our study suggests that genic and sub-genic intolerance are critical characteristics for interpreting the effects of variation in genes that influence epilepsy.
Keywords: ClinVar; Epi25; Louvain; epilepsy; epileptic encephalopathy; focal epilepsy; generalized epilepsy; intolerance; seizures; whole-exome sequencing.
Copyright © 2021 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
B.M.N. is a member of the scientific advisory board at Deep Genomics and RBNC Therapeutics, a member of the scientific advisory committee at Milken, and a consultant for Camp4 Therapeutics, Takeda Pharmaceutical, and Biogen. R.S.D. is a consultant for AstraZeneca. D.B.G. is a founder and shareholder in Praxis Precision Medicines, a shareholder in and member of the scientific advisor board for Apostle Inc., a shareholder in Q State – Biosciences, and a consultant for Gilead Sciences, AstraZeneca, and GoldFinch Bio.
Figures




References
-
- Aaberg K.M., Gunnes N., Bakken I.J., Lund Søraas C., Berntsen A., Magnus P., Lossius M.I., Stoltenberg C., Chin R., Surén P. Incidence and Prevalence of Childhood Epilepsy: A Nationwide Cohort Study. Pediatrics. 2017;139:e20163908. - PubMed
-
- Fisher R.S., Acevedo C., Arzimanoglou A., Bogacz A., Cross J.H., Elger C.E., Engel J., Jr., Forsgren L., French J.A., Glynn M. ILAE official report: a practical clinical definition of epilepsy. Epilepsia. 2014;55:475–482. - PubMed
-
- Ellis C.A., Petrovski S., Berkovic S.F. Epilepsy genetics: clinical impacts and biological insights. Lancet Neurol. 2020;19:93–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

