Genome-wide identification and analysis of class III peroxidases in Betula pendula
- PMID: 33932996
- PMCID: PMC8088069
- DOI: 10.1186/s12864-021-07622-1
Genome-wide identification and analysis of class III peroxidases in Betula pendula
Abstract
Background: Class III peroxidases (POD) proteins are widely present in the plant kingdom that are involved in a broad range of physiological processes including stress responses and lignin polymerization throughout the plant life cycle. At present, POD genes have been studied in Arabidopsis, rice, poplar, maize and Chinese pear, but there are no reports on the identification and function of POD gene family in Betula pendula.
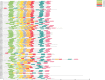
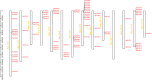
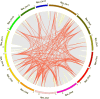
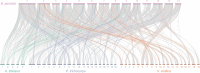
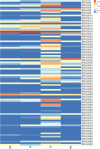
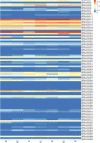
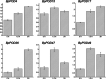
Results: We identified 90 nonredundant POD genes in Betula pendula. (designated BpPODs). According to phylogenetic relationships, these POD genes were classified into 12 groups. The BpPODs are distributed in different numbers on the 14 chromosomes, and some BpPODs were located sequentially in tandem on chromosomes. In addition, we analyzed the conserved domains of BpPOD proteins and found that they contain highly conserved motifs. We also investigated their expression patterns in different tissues, the results showed that some BpPODs might play an important role in xylem, leaf, root and flower. Furthermore, under low temperature conditions, some BpPODs showed different expression patterns at different times.
Conclusions: The research on the structure and function of the POD genes in Betula pendula plays a very important role in understanding the growth and development process and the molecular mechanism of stress resistance. These results lay the theoretical foundation for the genetic improvement of Betula pendula.
Keywords: Betula pendula; Chromosomal location; Class III peroxidases; Expression pattern; Phylogenetic analysis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Dunford HB, Stillman JS. On the function and mechanism of action of peroxidases. Coord Chem Rev. 1976;19(3):187–251. doi: 10.1016/S0010-8545(00)80316-1. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

