Biochemical mechanisms of period control within the mammalian circadian clock
- PMID: 33933351
- PMCID: PMC8551309
- DOI: 10.1016/j.semcdb.2021.04.012
Biochemical mechanisms of period control within the mammalian circadian clock
Abstract
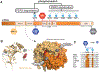
Genetically encoded biological clocks are found broadly throughout life on Earth, where they generate circadian (about a day) rhythms that synchronize physiology and behavior with the daily light/dark cycle. Although the genetic networks that give rise to circadian timing are now fairly well established, our understanding of how the proteins that constitute the molecular 'cogs' of this biological clock regulate the intrinsic timing, or period, of circadian rhythms has lagged behind. New studies probing the biochemical and structural basis of clock protein function are beginning to reveal how assemblies of dedicated clock proteins form and evolve through post-translational regulation to generate circadian rhythms. This review will highlight some recent advances providing important insight into the molecular mechanisms of period control in mammalian clocks with an emphasis on structural analyses related to CK1-dependent control of PER stability.
Keywords: Degradation; Dynamics; Feedback loop; Post-translational modifications; Proteins; Structure.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Figures


References
-
- Aschoff J: Circadian Rhythms in Man. Science 1965, 148(3676):1427–1432. - PubMed
-
- Bass J, Lazar MA: Circadian time signatures of fitness and disease. Science 2016, 354(6315):994–999. - PubMed
-
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM: Posttranslational mechanisms regulate the mammalian circadian clock. Cell 2001, 107(7):855–867. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

