Local Duplication of TIR-NBS-LRR Gene Marks Clubroot Resistance in Brassica napus cv. Tosca
- PMID: 33936130
- PMCID: PMC8082685
- DOI: 10.3389/fpls.2021.639631
Local Duplication of TIR-NBS-LRR Gene Marks Clubroot Resistance in Brassica napus cv. Tosca
Abstract
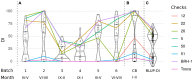
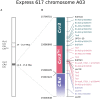
Clubroot, caused by Plasmodiophora brassicae infection, is a disease of growing importance in cruciferous crops, including oilseed rape (Brassica napus). The affected plants exhibit prominent galling of the roots that impairs their capacity for water and nutrient uptake, which leads to growth retardation, wilting, premature ripening, or death. Due to the scarcity of effective means of protection against the pathogen, breeding of resistant varieties remains a crucial component of disease control measures. The key aspect of the breeding process is the identification of genetic factors associated with variable response to the pathogen exposure. Although numerous clubroot resistance loci have been described in Brassica crops, continuous updates on the sources of resistance are necessary. Many of the resistance genes are pathotype-specific, moreover, resistance breakdowns have been reported. In this study, we characterize the clubroot resistance locus in the winter oilseed rape cultivar "Tosca." In a series of greenhouse experiments, we evaluate the disease severity of P. brassicae-challenged "Tosca"-derived population of doubled haploids, which we genotype with Brassica 60 K array and a selection of SSR/SCAR markers. We then construct a genetic map and narrow down the resistance locus to the 0.4 cM fragment on the A03 chromosome, corresponding to the region previously described as Crr3. Using Oxford Nanopore long-read genome resequencing and RNA-seq we review the composition of the locus and describe a duplication of TIR-NBS-LRR gene. Further, we explore the transcriptomic differences of the local genes between the clubroot resistant and susceptible, inoculated and control DH lines. We conclude that the duplicated TNL gene is a promising candidate for the resistance factor. This study provides valuable resources for clubroot resistance breeding programs and lays a foundation for further functional studies on clubroot resistance.
Keywords: Brassica napus; Oxford Nanopore; Plasmodiophora brassicae; QTL; RNA-Seq; TNL; duplication; resistance.
Copyright © 2021 Kopec, Mikolajczyk, Jajor, Perek, Nowakowska, Obermeier, Chawla, Korbas, Bartkowiak-Broda and Karlowski.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Clubroot resistance derived from the European Brassica napus cv. 'Tosca' is not effective against virulent Plasmodiophora brassicae isolates from Alberta, Canada.Sci Rep. 2021 Jul 14;11(1):14472. doi: 10.1038/s41598-021-93327-0. Sci Rep. 2021. PMID: 34262060 Free PMC article.
-
Clubroot resistance gene Rcr6 in Brassica nigra resides in a genomic region homologous to chromosome A08 in B. rapa.BMC Plant Biol. 2019 May 29;19(1):224. doi: 10.1186/s12870-019-1844-5. BMC Plant Biol. 2019. PMID: 31142280 Free PMC article.
-
HO-CR and HOLL-CR: new forms of winter oilseed rape (Brassica napus L.) with altered fatty acid composition and resistance to selected pathotypes of Plasmodiophora brassicae (clubroot).J Appl Genet. 2024 Sep;65(3):439-452. doi: 10.1007/s13353-024-00867-y. Epub 2024 Apr 19. J Appl Genet. 2024. PMID: 38637489 Free PMC article.
-
Plasmodiophora brassicae: a review of an emerging pathogen of the Canadian canola (Brassica napus) crop.Mol Plant Pathol. 2012 Feb;13(2):105-13. doi: 10.1111/j.1364-3703.2011.00729.x. Epub 2011 Jun 1. Mol Plant Pathol. 2012. PMID: 21726396 Free PMC article. Review.
-
The clubroot pathogen Plasmodiophora brassicae: A profile update.Mol Plant Pathol. 2023 Feb;24(2):89-106. doi: 10.1111/mpp.13283. Epub 2022 Nov 29. Mol Plant Pathol. 2023. PMID: 36448235 Free PMC article. Review.
Cited by
-
Integrating genome assembly, structural variation map construction and GWAS reveal the impact of SVs on agronomic traits of Brassica napus.Theor Appl Genet. 2025 Jul 26;138(8):191. doi: 10.1007/s00122-025-04977-x. Theor Appl Genet. 2025. PMID: 40715521
-
Pathotyping Systems and Pathotypes of Plasmodiophora brassicae-Navigating toward the Optimal Classification.Pathogens. 2024 Apr 11;13(4):313. doi: 10.3390/pathogens13040313. Pathogens. 2024. PMID: 38668268 Free PMC article. Review.
-
Lignin accumulation in cell wall plays a role in clubroot resistance.Front Plant Sci. 2024 Jul 23;15:1401265. doi: 10.3389/fpls.2024.1401265. eCollection 2024. Front Plant Sci. 2024. PMID: 39109069 Free PMC article.
-
Copy Number Variation among Resistance Genes Analogues in Brassica napus.Genes (Basel). 2022 Nov 4;13(11):2037. doi: 10.3390/genes13112037. Genes (Basel). 2022. PMID: 36360273 Free PMC article.
-
Dissection of a rapidly evolving wheat resistance gene cluster by long-read genome sequencing accelerated the cloning of Pm69.Plant Commun. 2024 Jan 8;5(1):100646. doi: 10.1016/j.xplc.2023.100646. Epub 2023 Jul 6. Plant Commun. 2024. PMID: 37415333 Free PMC article.
References
-
- Bates D., Mächler M., Bolker B., Walker S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67 1–48. 10.18637/jss.v067.i01 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

