Construction of a Machine Learning Dataset through Collaboration: The RSNA 2019 Brain CT Hemorrhage Challenge
- PMID: 33937827
- PMCID: PMC8082297
- DOI: 10.1148/ryai.2020190211
Construction of a Machine Learning Dataset through Collaboration: The RSNA 2019 Brain CT Hemorrhage Challenge
Erratum in
-
Erratum: Construction of a Machine Learning Dataset through Collaboration: The RSNA 2019 Brain CT Hemorrhage Challenge.Radiol Artif Intell. 2020 Jul 29;2(4):e209002. doi: 10.1148/ryai.2020209002. eCollection 2020 Jul. Radiol Artif Intell. 2020. PMID: 33939782 Free PMC article.
Abstract
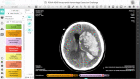
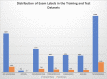
This dataset is composed of annotations of the five hemorrhage subtypes (subarachnoid, intraventricular, subdural, epidural, and intraparenchymal hemorrhage) typically encountered at brain CT.
2020 by the Radiological Society of North America, Inc.
Conflict of interest statement
Disclosures of Conflicts of Interest: A.E.F. disclosed no relevant relationships. L.M.P. Activities related to the present article: institution receives time-limited loaner access by OSU AI Lab to NVIDIA GPUs via Master Research Agreement between The Ohio State University and NVIDIA, no money transfer; time-limited access by OSU AI Lab to WIP postprocessing software via Master Research Agreement between OSU and Siemens Healthineers. No money transfer; unrestrictive support of OSU AI Lab by DeBartolo Family Funds. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. G.S. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: board member and shareholder of MD.ai but no money paid to author or MD.ai. Other relationships: disclosed no relevant relationships. S.S.H. disclosed no relevant relationships. J.K. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: author is consultant for INFOTECH, Soft; institution receives grants from Genentech and GE. Other relationships: disclosed no relevant relationships. R.B. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: previously employed by Roam Analytics (employment ended prior to working on this study); stockholder in Roam Analytics. Other relationships: disclosed no relevant relationships. J.T.M. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: consultant for Siemens (PAMA clinical decision support related); institution receives grants from GE (past and pending grants related to PTX detection); author may receive future royalties from GE related to PTX AI detector licensed to GE. Other relationships: disclosed no relevant relationships. A.S. Activities related to the present article: employee and shareholder in MD.ai. Activities not related to the present article: employee and shareholder in MD.ai. Other relationships: disclosed no relevant relationships. F.C.K. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: consultant for MD.ai; employed by Diagnósticos da América (DASA). Other relationships: disclosed no relevant relationships. M.P.L. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: consultant for and stockholder in Nines, SegMed, and Bunker Hill. Other relationships: disclosed no relevant relationships. G.C. disclosed no relevant relationships. L. Cala disclosed no relevant relationships. L. Coelho disclosed no relevant relationships. M.M. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: consultant for Cerebrotech Medical Systems. Other relationships: disclosed no relevant relationships. F.M. disclosed no relevant relationships. E.M. disclosed no relevant relationships. I.I. disclosed no relevant relationships. V.Z. disclosed no relevant relationships. O.M. disclosed no relevant relationships. C.L. disclosed no relevant relationships. L.S. disclosed no relevant relationships. D.J. disclosed no relevant relationships. A.A. disclosed no relevant relationships. R.K.L. disclosed no relevant relationships. J.N. disclosed no relevant relationships.
Figures



References
-
- Cordonnier C , Demchuk A , Ziai W , Anderson CS . Intracerebral haemorrhage: current approaches to acute management . Lancet 2018. ; 392 ( 10154 ): 1257 – 1268 . - PubMed
-
- Majumdar A , Brattain L , Telfer B , Farris C , Scalera J . Detecting Intracranial Hemorrhage with Deep Learning . Conf Proc IEEE Eng Med Biol Soc 2018. ; 2018 : 583 – 587 . - PubMed
-
- Ginat DT . Analysis of head CT scans flagged by deep learning software for acute intracranial hemorrhage . Neuroradiology 2020. ; 62 ( 3 ): 335 – 340 . - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

