MRI Manufacturer Shift and Adaptation: Increasing the Generalizability of Deep Learning Segmentation for MR Images Acquired with Different Scanners
- PMID: 33937833
- PMCID: PMC8082399
- DOI: 10.1148/ryai.2020190195
MRI Manufacturer Shift and Adaptation: Increasing the Generalizability of Deep Learning Segmentation for MR Images Acquired with Different Scanners
Abstract
Purpose: To quantitatively evaluate the generalizability of a deep learning segmentation tool to MRI data from scanners of different MRI manufacturers and to improve the cross-manufacturer performance by using a manufacturer-adaptation strategy.
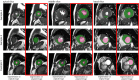
Materials and methods: This retrospective study included 150 cine MRI datasets from three MRI manufacturers, acquired between 2017 and 2018 (n = 50 for manufacturer 1, manufacturer 2, and manufacturer 3). Three convolutional neural networks (CNNs) were trained to segment the left ventricle (LV), using datasets exclusively from images from a single manufacturer. A generative adversarial network (GAN) was trained to adapt the input image before segmentation. The LV segmentation performance, end-diastolic volume (EDV), end-systolic volume (ESV), LV mass, and LV ejection fraction (LVEF) were evaluated before and after manufacturer adaptation. Paired Wilcoxon signed rank tests were performed.
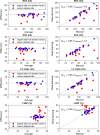
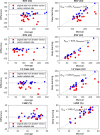
Results: The segmentation CNNs exhibited a significant performance drop when applied to datasets from different manufacturers (Dice reduced from 89.7% ± 2.3 [standard deviation] to 68.7% ± 10.8, P < .05, from 90.6% ± 2.1 to 59.5% ± 13.3, P < .05, from 89.2% ± 2.3 to 64.1% ± 12.0, P < .05, for manufacturer 1, 2, and 3, respectively). After manufacturer adaptation, the segmentation performance was significantly improved (from 68.7% ± 10.8 to 84.3% ± 6.2, P < .05, from 72.4% ± 10.2 to 85.7% ± 6.5, P < .05, for manufacturer 2 and 3, respectively). Quantitative LV function parameters were also significantly improved. For LVEF, the manufacturer adaptation increased the Pearson correlation from 0.005 to 0.89 for manufacturer 2 and from 0.77 to 0.94 for manufacturer 3.
Conclusion: A segmentation CNN well trained on datasets from one MRI manufacturer may not generalize well to datasets from other manufacturers. The proposed manufacturer adaptation can largely improve the generalizability of a deep learning segmentation tool without additional annotation.Supplemental material is available for this article.© RSNA, 2020.
2020 by the Radiological Society of North America, Inc.
Conflict of interest statement
Disclosures of Conflicts of Interest: W.Y. Activities related to the present article: institution received money from National Key Research and Development Program of China under Grant 2018YFC0116303. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. L.H. disclosed no relevant relationships. L.X. disclosed no relevant relationships. S.G. disclosed no relevant relationships. F.Y. disclosed no relevant relationships. Y.W. Activities related to the present article: institution received money from National Key Research and Development Program of China under Grant 2018YFC0116303. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. Q.T. disclosed no relevant relationships.
Figures


References
-
- McRobbie DW, Moore EA, Graves MJ, Prince MR. MRI from Picture to Proton. Cambridge Core. 2006. http://cambridge.org/core/books/mri-from-picture-to-proton/3ADC814FF8FC6.... Accessed October 10, 2019.
-
- Lakhani P, Prater AB, Hutson RK, et al. . Machine Learning in Radiology: Applications Beyond Image Interpretation. J Am Coll Radiol 2018;15(2):350–359. - PubMed
-
- Tao Q, Yan W, Wang Y, et al. . Deep Learning-based Method for Fully Automatic Quantification of Left Ventricle Function from Cine MR Images: A Multivendor, Multicenter Study. Radiology 2019;290(1):81–88. - PubMed
-
- Kurata Y, Nishio M, Kido A, et al. . Automatic segmentation of the uterus on MRI using a convolutional neural network. Comput Biol Med 2019;114:103438. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

