The multi-functionality of UHRF1: epigenome maintenance and preservation of genome integrity
- PMID: 33939809
- PMCID: PMC8216287
- DOI: 10.1093/nar/gkab293
The multi-functionality of UHRF1: epigenome maintenance and preservation of genome integrity
Abstract
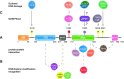
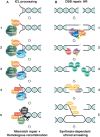
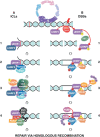
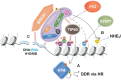
During S phase, the cooperation between the macromolecular complexes regulating DNA synthesis, epigenetic information maintenance and DNA repair is advantageous for cells, as they can rapidly detect DNA damage and initiate the DNA damage response (DDR). UHRF1 is a fundamental epigenetic regulator; its ability to coordinate DNA methylation and histone code is unique across proteomes of different species. Recently, UHRF1's role in DNA damage repair has been explored and recognized to be as important as its role in maintaining the epigenome. UHRF1 is a sensor for interstrand crosslinks and a determinant for the switch towards homologous recombination in the repair of double-strand breaks; its loss results in enhanced sensitivity to DNA damage. These functions are finely regulated by specific post-translational modifications and are mediated by the SRA domain, which binds to damaged DNA, and the RING domain. Here, we review recent studies on the role of UHRF1 in DDR focusing on how it recognizes DNA damage and cooperates with other proteins in its repair. We then discuss how UHRF1's epigenetic abilities in reading and writing histone modifications, or its interactions with ncRNAs, could interlace with its role in DDR.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Muto M., Kanari Y., Kubo E., Takabe T., Kurihara T., Fujimori A., Tatsumi K.. Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J. Biol. Chem. 2002; 277:34549–34555. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

