Informing NMR experiments with molecular dynamics simulations to characterize the dominant activated state of the KcsA ion channel
- PMID: 33940802
- PMCID: PMC9250420
- DOI: 10.1063/5.0040649
Informing NMR experiments with molecular dynamics simulations to characterize the dominant activated state of the KcsA ion channel
Abstract
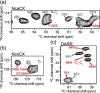
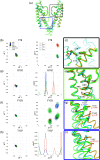
As the first potassium channel with an x-ray structure determined, and given its homology to eukaryotic channels, the pH-gated prokaryotic channel KcsA has been extensively studied. Nevertheless, questions related, in particular, to the allosteric coupling between its gates remain open. The many currently available x-ray crystallography structures appear to correspond to various stages of activation and inactivation, offering insights into the molecular basis of these mechanisms. Since these studies have required mutations, complexation with antibodies, and substitution of detergents in place of lipids, examining the channel under more native conditions is desirable. Solid-state nuclear magnetic resonance (SSNMR) can be used to study the wild-type protein under activating conditions (low pH), at room temperature, and in bacteriomimetic liposomes. In this work, we sought to structurally assign the activated state present in SSNMR experiments. We used a combination of molecular dynamics (MD) simulations, chemical shift prediction algorithms, and Bayesian inference techniques to determine which of the most plausible x-ray structures resolved to date best represents the activated state captured in SSNMR. We first identified specific nuclei with simulated NMR chemical shifts that differed significantly when comparing partially open vs fully open ensembles from MD simulations. The simulated NMR chemical shifts for those specific nuclei were then compared to experimental ones, revealing that the simulation of the partially open state was in good agreement with the SSNMR data. Nuclei that discriminate effectively between partially and fully open states belong to residues spread over the sequence and provide a molecular level description of the conformational change.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

