Genomic analysis of novel Yarrowia-like yeast symbionts associated with the carrion-feeding burying beetle Nicrophorus vespilloides
- PMID: 33941076
- PMCID: PMC8091737
- DOI: 10.1186/s12864-021-07597-z
Genomic analysis of novel Yarrowia-like yeast symbionts associated with the carrion-feeding burying beetle Nicrophorus vespilloides
Abstract
Background: Mutualistic interactions with microbes can help insects adapt to extreme environments and unusual diets. An intriguing example is the burying beetle Nicrophorus vespilloides, which feeds and reproduces on small vertebrate carcasses. Its fungal microbiome is dominated by yeasts that potentially facilitate carcass utilization by producing digestive enzymes, eliminating cadaver-associated toxic volatiles (that would otherwise attract competitors), and releasing antimicrobials to sanitize the microenvironment. Some of these yeasts are closely related to the biotechnologically important species Yarrowia lipolytica.
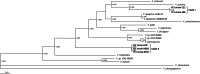
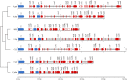
Results: To investigate the roles of these Yarrowia-like yeast (YLY) strains in more detail, we selected five strains from two different phylogenetic clades for third-generation sequencing and genome analysis. The first clade, represented by strain B02, has a 20-Mb genome containing ~ 6400 predicted protein-coding genes. The second clade, represented by strain C11, has a 25-Mb genome containing ~ 6300 predicted protein-coding genes, and extensive intraspecific variability within the ITS-D1/D2 rDNA region commonly used for species assignments. Phenotypic microarray analysis revealed that both YLY strains were able to utilize a diverse range of carbon and nitrogen sources (including microbial metabolites associated with putrefaction), and can grow in environments with extreme pH and salt concentrations.
Conclusions: The genomic characterization of five yeast strains isolated from N. vespilloides resulted in the identification of strains potentially representing new YLY species. Given their abundance in the beetle hindgut, and dominant growth on beetle-prepared carcasses, the analysis of these strains has revealed the genetic basis of a potential symbiotic relationship between yeasts and burying beetles that facilitates carcass digestion and preservation.
Keywords: Carrion beetles; Detoxification; Digestion; Ephemeral resources; Metabolic profiling; Nicrophorus vespilloides; Transcriptomics; Yarrowia; rDNA variability.
Conflict of interest statement
All authors declare that they have no competing interests.
Figures





References
-
- Mueller UG, Rehner SA, Schultz TR. The evolution of agriculture in ants. Science. 1998;281(5385):2034–2038. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

