Whole-genome sequencing analysis of semi-supercentenarians
- PMID: 33941312
- PMCID: PMC8096429
- DOI: 10.7554/eLife.57849
Whole-genome sequencing analysis of semi-supercentenarians
Abstract
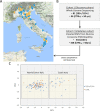
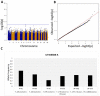
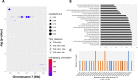
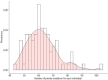
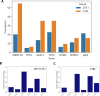
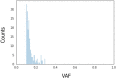
Extreme longevity is the paradigm of healthy aging as individuals who reached the extreme decades of human life avoided or largely postponed all major age-related diseases. In this study, we sequenced at high coverage (90X) the whole genome of 81 semi-supercentenarians and supercentenarians [105+/110+] (mean age: 106.6 ± 1.6) and of 36 healthy unrelated geographically matched controls (mean age 68.0 ± 5.9) recruited in Italy. The results showed that 105+/110+ are characterized by a peculiar genetic background associated with efficient DNA repair mechanisms, as evidenced by both germline data (common and rare variants) and somatic mutations patterns (lower mutation load if compared to younger healthy controls). Results were replicated in a second independent cohort of 333 Italian centenarians and 358 geographically matched controls. The genetics of 105+/110+ identified DNA repair and clonal haematopoiesis as crucial players for healthy aging and for the protection from cardiovascular events.
Keywords: ageing; clonal hematopoiesis; genetics; genomics; geroscience; human; longevity; semi-supercentenarians; sequencing.
© 2021, Garagnani et al.
Conflict of interest statement
PG, MD, CP, EM, KK, VI, LX, DM, BA, MC, EF, DM, BN, SS, DL, DP, GC, CS, GP, FD, PD, LB, NM, DG, OO, CG, CF No competing interests declared, JM, AV, JC, FR, SC, PD is affiliated with Nestlé Research, Société des Produits Nestlé SA. The author has no other competing interests to declare. MB s affiliated with Nestlé Research, Société des Produits Nestlé SA. The author has no other competing interests to declare. AF is affiliated with Menarini Silicon Biosystems SpA. The author has no other competing interests to declare.
Figures



References
-
- Andersen SL, Sebastiani P, Dworkis DA, Feldman L, Perls TT. Health span approximates life span among many supercentenarians: compression of morbidity at the approximate limit of life span. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 2012;67A:395–405. doi: 10.1093/gerona/glr223. - DOI - PMC - PubMed
-
- Beekman M, Nederstigt C, Suchiman HE, Kremer D, van der Breggen R, Lakenberg N, Alemayehu WG, de Craen AJ, Westendorp RG, Boomsma DI, de Geus EJ, Houwing-Duistermaat JJ, Heijmans BT, Slagboom PE. Genome-wide association study (GWAS)-identified disease risk alleles do not compromise human longevity. PNAS. 2010;107:18046–18049. doi: 10.1073/pnas.1003540107. - DOI - PMC - PubMed
-
- Bonafè M, Olivieri F, Mari D, Baggio G, Mattace R, Sansoni P, De Benedictis G, De Luca M, Bertolini S, Barbi C, Monti D, Franceschi C. p53 variants predisposing to Cancer are present in healthy centenarians. The American Journal of Human Genetics. 1999;64:292–294. doi: 10.1086/302196. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

