Protein structure-based gene expression signatures
- PMID: 33941686
- PMCID: PMC8126868
- DOI: 10.1073/pnas.2014866118
Protein structure-based gene expression signatures
Abstract
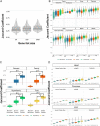
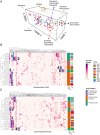
Gene expression signatures (GES) connect phenotypes to differential messenger RNA (mRNA) expression of genes, providing a powerful approach to define cellular identity, function, and the effects of perturbations. The use of GES has suffered from vague assessment criteria and limited reproducibility. Because the structure of proteins defines the functional capability of genes, we hypothesized that enrichment of structural features could be a generalizable representation of gene sets. We derive structural gene expression signatures (sGES) using features from multiple levels of protein structure (e.g., domain and fold) encoded by the mRNAs in GES. Comprehensive analyses of data from the Genotype-Tissue Expression Project (GTEx), the all RNA-seq and ChIP-seq sample and signature search (ARCHS4) database, and mRNA expression of drug effects on cardiomyocytes show that sGES are useful for characterizing biological phenomena. sGES enable phenotypic characterization across experimental platforms, facilitates interoperability of expression datasets, and describe drug action on cells.
Keywords: gene expression signatures; reproducibility; structural bioinformatics.
Conflict of interest statement
Competing interest statement: R.R. and A.S. are co-founders of Aichemy Inc.
Figures





References
-
- Li M., et al. ., HiFreSP: A novel high-frequency sub-pathway mining approach to identify robust prognostic gene signatures. Brief. Bioinform. 21, 1411–1424 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

