Molecular and clinical determinants of response and resistance to rucaparib for recurrent ovarian cancer treatment in ARIEL2 (Parts 1 and 2)
- PMID: 33941784
- PMCID: PMC8093258
- DOI: 10.1038/s41467-021-22582-6
Molecular and clinical determinants of response and resistance to rucaparib for recurrent ovarian cancer treatment in ARIEL2 (Parts 1 and 2)
Abstract
ARIEL2 (NCT01891344) is a single-arm, open-label phase 2 study of the PARP inhibitor (PARPi) rucaparib in relapsed high-grade ovarian carcinoma. In this post hoc exploratory biomarker analysis of pre- and post-platinum ARIEL2 samples, RAD51C and RAD51D mutations and high-level BRCA1 promoter methylation predict response to rucaparib, similar to BRCA1/BRCA2 mutations. BRCA1 methylation loss may be a major cross-resistance mechanism to platinum and PARPi. Genomic scars associated with homologous recombination deficiency are irreversible, persisting even as platinum resistance develops, and therefore are predictive of rucaparib response only in platinum-sensitive disease. The RAS, AKT, and cell cycle pathways may be additional modulators of PARPi sensitivity.
Conflict of interest statement
E.M.S. has received funding for clinical trials from Clovis Oncology and TESARO (paid to her institution). T.T.K., E.D., L.-T.V., S.G., L. Maloney, H.G., T.H., and K.K.L. are employees of Clovis Oncology and may own stock or have stock options in that company. A.M.O. has served on advisory boards for Clovis Oncology, Amgen, Immunovaccine, and Verastem; received support for travel or accommodation from AstraZeneca; and received honoraria from WebRx. A.V.T. has served on an advisory board for and received grants from AstraZeneca. I.R.-C. has served on advisory boards for Clovis Oncology, AstraZeneca, Genmab/Seattle Genetics, ImmunoGen, Merck Sharpe Dohme, PharmaMar, Roche, and Tesaro/GlaxoSmithKline and received support for travel or accommodation from AstraZeneca, GlaxoSmithKline, and Roche. A.O. has served on advisory boards for Clovis Oncology, AstraZeneca, ImmunoGen, Genmab/Seattle Genetics, PharmaMar, Roche, and Tesaro and received support for travel or accommodation from AstraZeneca, PharmaMar, Roche, and Tesaro. R.L.C. reports grants from Clovis Oncology, AbbVie, AstraZeneca, Esperance, Janssen, Merck, Millennium, OncoMed, and Roche/Genentech and has served as an advisor to Clovis Oncology, AbbVie, AstraZeneca, Bayer, Esperance, GamaMabs, Genmab, Gradalis, Janssen, Millennium, Merck, OncoMed, Pfizer, Roche/Genentech, and Tesaro. C.A. has served on steering committees for Clovis Oncology and Mateon Therapeutics; served on advisory boards for Clovis Oncology, Bayer, Cerulean Pharma, Tesaro, and VentiRx, and received honoraria from Clovis Oncology, Bayer, Cerulean Pharma, Mateon Therapeutics, Tesaro, and VentiRx. G.E.K. has received research funding (to the University of California outside the scope of this work) from Lilly, Merck, and Pfizer, and received honorarium from Clovis Oncology, AstraZeneca, and Tesaro. D.M.O. has served on advisory boards for Clovis Oncology, AstraZeneca, Gynecologic Oncology Group, Janssen, Myriad, and Tesaro; has served on steering committees for Clovis Oncology, Amgen, and ImmunoGen; has served as a consultant to AbbVie, Ambry, AstraZeneca, Health Analytics, and Tesaro, and his institution has received research support from Clovis Oncology, Agenus, Ajinomoto, Array BioPharma, AstraZeneca, Bristol-Myers Squibb, ERGOMED Clinical Research, Exelixis, Genentech, GlaxoSmithKline, Gynecologic Oncology Group, ImmunoGen, INC Research, inVentiv Health Clinical, Janssen Research and Development, Ludwig Institute for Cancer Research, Novartis Pharmaceuticals, PRA International, Regeneron Pharmaceuticals, Serono, Stemcentrx, Tesaro, and TRACON Pharmaceuticals. A.L. has served on advisory boards for Clovis Oncology, Ability, AstraZeneca, Biocad, Merck Serono, MSD, Pfizer, PharmaMar, Seattle Genetics, and Tesaro PharmaMar; reports institutional research grant support from GamaMabs, Inivata, Merus, and Sanofi, and reports boarding and travel expenses for congress activities from Clovis Oncology, AstraZeneca, Roche, and Tesaro. D.P. has served on advisory boards for AstraZeneca and GlaxoSmithKline, and received support for travel from AstraZeneca. S.W. has served on advisory boards for AstraZeneca and Roche; reports institutional research support from Clovis Oncology, AstraZeneca, Merck, Regeneron, and Tesaro, and has received honoraria from AstraZeneca. P.G. has served on an advisory board for AstraZeneca, GSK, and Merck, and received support for travel expenses for congress activities from AstraZeneca. R.S.K. has served on advisory boards for Clovis Oncology, Roche, and Tesaro. O.D. has served on advisory boards for Clovis Oncology, IMV, Tesaro, and Merck, and has served on the speaker bureau for AstraZeneca and Tesaro. J.A.E. and D.I.L. are employees of Foundation Medicine, Inc., which is a wholly-owned subsidiary of Roche, and may own stock or have stock options in Roche. A.D. has served on advisory boards for AstraZeneca Australia and has received travel support from Bio-Rad. C.L.S. has served on advisory boards for Clovis Oncology, AstraZeneca, Takeda, Roche Australia, MSD, Eisai Co., has received travel support from AstraZeneca, has received in kind research support from Clovis Oncology, Roche, Beigene and Funded research support from Sierra oncology and Eisai Co. I.A.M. has served on advisory boards for Clovis Oncology, AstraZeneca, Roche, Takeda, and Tesaro and receives institutional funding from AstraZeneca. The remaining authors declare no competing interests.
Figures
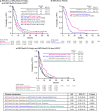


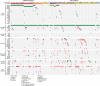
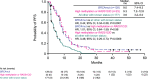
References
-
- Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5:1137–1154. doi: 10.1158/2159-8290.CD-15-0714. - DOI - PMC - PubMed
-
- Ledermann JA, et al. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: an updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Oncol. 2016;17:1579–1589. doi: 10.1016/S1470-2045(16)30376-X. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

