The PI3K pathway as a therapeutic intervention point in inflammatory bowel disease
- PMID: 33942546
- PMCID: PMC8342202
- DOI: 10.1002/iid3.435
The PI3K pathway as a therapeutic intervention point in inflammatory bowel disease
Abstract
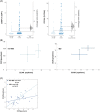
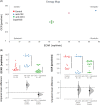
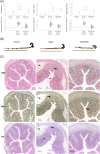
With glucose being the preferred source of energy in activated T cells, targeting glycolysis has become an attractive therapeutic intervention point for chronic inflammatory bowel diseases (IBD). The switch to glycolysis is mediated by phosphoinositide-3-kinases (PI3K) which relay signals from surface receptors to the AKT pathway. We first confirmed by analysis of the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) that metabolism is shifted towards glycolysis in IBD patients as compared to non-IBD donors. In contrast to non-IBD donors, OCR correlated with ECAR (IBD: cor = 0.79, p = 2E-10; non-IBD: cor = 0.37, p = n.s.), in IBD patients. Second, we tested the PI3K inhibitor copanlisib as a potential therapeutic. Ex vivo, copanlisib suppressed the ECAR significantly in T cells activated by anti-CD3 antibodies and significantly decreased ECAR rates in the presence of copanlisib (anti-CD3: 58.24 ± 29.06; copanlisib: 43.16 ± 20.23, p < .000. In addition, copanlisib impaired the activation of CD4+ CD25+ T cells (anti-CD3: 42.15 ± 21.46; anti-CD3 + copanlisib: 26.06 ± 21.82 p = .013) and the secretion of cytokines (IFNγ: anti-CD3: 6332.0 ± 5707.61 pmol/ml; anti-CD3 + copanlisib: 6332.0 ± 5707.61, p = .018). In vivo, copanlisib significantly improved the histological scores (ethanol: 8.5 ± 3.81; copanlisib: 4.57 ± 2.82, p = .006) in the NSG-UC mouse model. Orthogonal partial least square analysis confirmed the efficacy of copanlisib. These data suggest that the PI3K pathway provides an attractive therapeutic intervention point in IBD for patients in relapse. Targeting metabolic pathways have the potential to develop phase dependent therapies.
Keywords: NSG-UC mouse model; PI3K; copanlisib; immune-metabolism; inflammatory bowel disease; ulcerative colitis.
© 2021 The Authors. Immunity, Inflammation and Disease published by John Wiley & Sons Ltd.
Conflict of interest statement
Paula Winkelmann, Anna‐Lena Unterweger, and Diya Khullar are supported by Shaw Research. Roswitha Gropp has a consulting agreement with Shaw Research.
Figures



References
-
- Head KA, Jurenka JS. Inflammatory bowel disease part I: ulcerative colitis–pathophysiology and conventional and alternative treatment options. Altern Med Rev. 2003;8:247‐283. - PubMed
-
- Health NIo. Ulcerative Colitis. Genetics Home Reference: Lister Hill National Center for Biomedical Communications, National Institutes of Health, Department of Health & Human Services, 2019.
-
- Kornbluth A, Sachar DB. Ulcerative colitis practice guidelines in adults: American college of gastroenterology, practice parameters committee. Am J Gastroenterol. 2010;105:501‐523. - PubMed
-
- Conrad K, Roggenbuck D, Laass MW. Diagnosis and classification of ulcerative colitis. Autoimmun Rev. 2014;13:463‐466. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

