Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3
- PMID: 33944776
- PMCID: PMC8096432
- DOI: 10.7554/eLife.65088
Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3
Abstract
Culture-independent analyses of microbial communities have progressed dramatically in the last decade, particularly due to advances in methods for biological profiling via shotgun metagenomics. Opportunities for improvement continue to accelerate, with greater access to multi-omics, microbial reference genomes, and strain-level diversity. To leverage these, we present bioBakery 3, a set of integrated, improved methods for taxonomic, strain-level, functional, and phylogenetic profiling of metagenomes newly developed to build on the largest set of reference sequences now available. Compared to current alternatives, MetaPhlAn 3 increases the accuracy of taxonomic profiling, and HUMAnN 3 improves that of functional potential and activity. These methods detected novel disease-microbiome links in applications to CRC (1262 metagenomes) and IBD (1635 metagenomes and 817 metatranscriptomes). Strain-level profiling of an additional 4077 metagenomes with StrainPhlAn 3 and PanPhlAn 3 unraveled the phylogenetic and functional structure of the common gut microbe Ruminococcus bromii, previously described by only 15 isolate genomes. With open-source implementations and cloud-deployable reproducible workflows, the bioBakery 3 platform can help researchers deepen the resolution, scale, and accuracy of multi-omic profiling for microbial community studies.
Keywords: computational analysis; computational biology; human; infectious disease; metagenomics; microbial genomics; microbiology; microbiome; systems biology.
© 2021, Beghini et al.
Conflict of interest statement
FB, LM, AB, LD, FA, SM, AM, PM, MS, AT, MV, GW, YZ, MZ, CH, EF, NS No competing interests declared
Figures







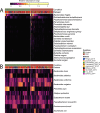





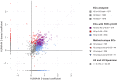


References
-
- Almeida A, Nayfach S, Boland M, Strozzi F, Beracochea M, Shi ZJ, Pollard KS, Sakharova E, Parks DH, Hugenholtz P, Segata N, Kyrpides NC, Finn RD. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nature Biotechnology. 2021;39:105–114. doi: 10.1038/s41587-020-0603-3. - DOI - PMC - PubMed
-
- Andrews S O. FastQC: A Quality Control Tool for High Throughput Sequence Data 2010
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology the gene ontology consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

