Dead or alive: microbial viability treatment reveals both active and inactive bacterial constituents in the fish gut microbiota
- PMID: 33945191
- PMCID: PMC8596808
- DOI: 10.1111/jam.15113
Dead or alive: microbial viability treatment reveals both active and inactive bacterial constituents in the fish gut microbiota
Abstract
Aims: This study evaluated the microbial viability of fish gut microbiota in both digesta (faecal) and mucosal samples using a modified propidium monoazide (PMA) protocol, followed by 16S ribosomal RNA (rRNA) gene sequencing.
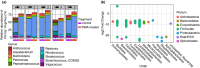
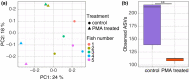
Methods and results: Digesta and gut mucosal samples from farmed yellowtail kingfish (Seriola lalandi) were collected and a modified PMA treatment was applied prior to DNA extraction to differentiate both active and nonviable microbial cells in the samples. All samples were then sequenced using a standard 16S rRNA approach. The digesta and mucosal samples contained significantly different bacterial communities, with a higher diversity observed in digesta samples. In addition, PMA treatment significantly reduced the microbial diversity and richness of digesta and mucosal samples and depleted bacterial constituents typically considered to be important within fish, such as Lactobacillales and Clostridales taxa.
Conclusions: These findings suggest that important bacterial members may not be active in the fish gut microbiota. In particular, several beneficial lactic acid bacteria (LAB) were identified as nonviable bacterial cells, potentially influencing the functional potential of the fish microbiota.
Significance and impacts of the study: Standardizing the methods for characterizing the fish microbiota are paramount in order to compare studies. In this study, we showed that both sample type and PMA treatment influence the bacterial communities found in the fish gut microbiota. Our findings also suggest that several microbes previously described in the fish gut may not be active constituents. As a result, these factors should be considered in future studies to better evaluate the active bacterial communities associated with the host.
Keywords: 16S rRNA; PMA; fish; gut; microbiota; viability; yellowtail kingfish.
© 2021 The Authors. Journal of Applied Microbiology published by John Wiley & Sons Ltd on behalf of Society for Applied Microbiology.
Conflict of interest statement
No conflict of interest declared.
Figures




References
-
- Anderson, M.J. (2001) A new method for non‐parametric multivariate analysis of variance. Austral Ecol 26, 32–46.
-
- Banerjee, G. and Ray, A.K. (2017) Bacterial symbiosis in the fish gut and its role in health and metabolism. Symbiosis 72, 1–11.
-
- de Bruijn, I. , Liu, Y.Y. , Wiegertjes, G.F. and Raaijmakers, J.M. (2018) Exploring fish microbial communities to mitigate emerging diseases in aquaculture. FEMS Microbiol Ecol 94, 12. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

