Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII
- PMID: 33945229
- PMCID: PMC8095314
- DOI: 10.1002/open.202100042
Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII
Abstract
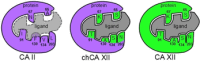
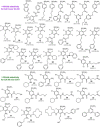
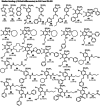
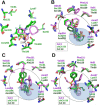
A key part of the optimization of small molecules in pharmaceutical inhibitor development is to vary the molecular design to enhance complementarity of chemical features of the compound with the positioning of amino acids in the active site of a target enzyme. Typically this involves iterations of synthesis, to modify the compound, and biophysical assay, to assess the outcomes. Selective targeting of the anti-cancer carbonic anhydrase isoform XII (CA XII), this process is challenging because the overall fold is very similar across the twelve CA isoforms. To enhance drug development for CA XII we used a reverse engineering approach where mutation of the key six amino acids in the active site of human CA XII into the CA II isoform was performed to provide a protein chimera (chCA XII) which is amenable to structure-based compound optimization. Through determination of structural detail and affinity measurement of the interaction with over 60 compounds we observed that the compounds that bound CA XII more strongly than CA II, switched their preference and bound more strongly to the engineered chimera, chCA XII, based on CA II, but containing the 6 key amino acids from CA XII, behaved as CA XII in its compound recognition profile. The structures of the compounds in the chimeric active site also resembled those determined for complexes with CA XII, hence validating this protein engineering approach in the development of new inhibitors.
Keywords: X-ray diffraction; drug design; enzyme models; fluorescence spectroscopy; sulfonamides.
© 2021 The Authors. Published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

