Ranking microbial metabolomic and genomic links in the NPLinker framework using complementary scoring functions
- PMID: 33945539
- PMCID: PMC8130963
- DOI: 10.1371/journal.pcbi.1008920
Ranking microbial metabolomic and genomic links in the NPLinker framework using complementary scoring functions
Abstract
Specialised metabolites from microbial sources are well-known for their wide range of biomedical applications, particularly as antibiotics. When mining paired genomic and metabolomic data sets for novel specialised metabolites, establishing links between Biosynthetic Gene Clusters (BGCs) and metabolites represents a promising way of finding such novel chemistry. However, due to the lack of detailed biosynthetic knowledge for the majority of predicted BGCs, and the large number of possible combinations, this is not a simple task. This problem is becoming ever more pressing with the increased availability of paired omics data sets. Current tools are not effective at identifying valid links automatically, and manual verification is a considerable bottleneck in natural product research. We demonstrate that using multiple link-scoring functions together makes it easier to prioritise true links relative to others. Based on standardising a commonly used score, we introduce a new, more effective score, and introduce a novel score using an Input-Output Kernel Regression approach. Finally, we present NPLinker, a software framework to link genomic and metabolomic data. Results are verified using publicly available data sets that include validated links.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

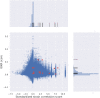
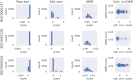
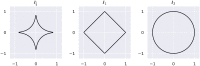
Similar articles
-
Enhanced correlation-based linking of biosynthetic gene clusters to their metabolic products through chemical class matching.Microbiome. 2023 Jan 23;11(1):13. doi: 10.1186/s40168-022-01444-3. Microbiome. 2023. PMID: 36691088 Free PMC article.
-
Recent development of antiSMASH and other computational approaches to mine secondary metabolite biosynthetic gene clusters.Brief Bioinform. 2019 Jul 19;20(4):1103-1113. doi: 10.1093/bib/bbx146. Brief Bioinform. 2019. PMID: 29112695 Free PMC article.
-
MIBiG 2.0: a repository for biosynthetic gene clusters of known function.Nucleic Acids Res. 2020 Jan 8;48(D1):D454-D458. doi: 10.1093/nar/gkz882. Nucleic Acids Res. 2020. PMID: 31612915 Free PMC article.
-
Computational strategies for genome-based natural product discovery and engineering in fungi.Fungal Genet Biol. 2016 Apr;89:29-36. doi: 10.1016/j.fgb.2016.01.006. Epub 2016 Jan 13. Fungal Genet Biol. 2016. PMID: 26775250 Review.
-
Linking genomics and metabolomics to chart specialized metabolic diversity.Chem Soc Rev. 2020 Jun 7;49(11):3297-3314. doi: 10.1039/d0cs00162g. Epub 2020 May 12. Chem Soc Rev. 2020. PMID: 32393943 Review.
Cited by
-
Enhanced correlation-based linking of biosynthetic gene clusters to their metabolic products through chemical class matching.Microbiome. 2023 Jan 23;11(1):13. doi: 10.1186/s40168-022-01444-3. Microbiome. 2023. PMID: 36691088 Free PMC article.
-
Primed for Discovery.Biochemistry. 2024 Nov 5;63(21):2705-2713. doi: 10.1021/acs.biochem.4c00464. Epub 2024 Oct 15. Biochemistry. 2024. PMID: 39497571 Free PMC article. Review.
-
antiSMASH 8.0: extended gene cluster detection capabilities and analyses of chemistry, enzymology, and regulation.Nucleic Acids Res. 2025 Jul 7;53(W1):W32-W38. doi: 10.1093/nar/gkaf334. Nucleic Acids Res. 2025. PMID: 40276974 Free PMC article.
-
Correlative metabologenomics of 110 fungi reveals metabolite-gene cluster pairs.Nat Chem Biol. 2023 Jul;19(7):846-854. doi: 10.1038/s41589-023-01276-8. Epub 2023 Mar 6. Nat Chem Biol. 2023. PMID: 36879060 Free PMC article.
-
Mining genomes to illuminate the specialized chemistry of life.Nat Rev Genet. 2021 Sep;22(9):553-571. doi: 10.1038/s41576-021-00363-7. Epub 2021 Jun 3. Nat Rev Genet. 2021. PMID: 34083778 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

