Targeting microRNA-mediated gene repression limits adipogenic conversion of skeletal muscle mesenchymal stromal cells
- PMID: 33945794
- PMCID: PMC8254802
- DOI: 10.1016/j.stem.2021.04.008
Targeting microRNA-mediated gene repression limits adipogenic conversion of skeletal muscle mesenchymal stromal cells
Abstract
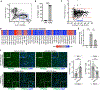
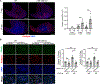
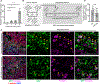
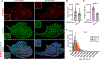
Intramuscular fatty deposits, which are seen in muscular dystrophies and with aging, negatively affect muscle function. The cells of origin of adipocytes constituting these fatty deposits are mesenchymal stromal cells, fibroadipogenic progenitors (FAPs). We uncover a molecular fate switch, involving miR-206 and the transcription factor Runx1, that controls FAP differentiation to adipocytes. Mice deficient in miR-206 exhibit increased adipogenesis following muscle injury. Adipogenic differentiation of FAPs is abrogated by miR-206 mimics. Using a labeled microRNA (miRNA) pull-down and sequencing (LAMP-seq), we identified Runx1 as a miR-206 target, with miR-206 repressing Runx1 translation. In the absence of miR-206 in FAPs, Runx1 occupancy near transcriptional start sites of adipogenic genes and expression of these genes increase. We demonstrate that miR-206 mimicry in vivo limits intramuscular fatty infiltration. Our results provide insight into the underlying molecular mechanisms of FAP fate determination and formation of harmful fatty deposits in skeletal muscle.
Keywords: adipogenesis; aging; cell fate determination; fatty infiltration; fibroadipogenic progenitor; mesenchymal stromal cell; muscular dystrophy; skeletal muscle; stem cell.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Bray NL, Pimentel H, Melsted P, Pachter L, 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

