DNA Methylation Change Profiling of Colorectal Disease: Screening towards Clinical Use
- PMID: 33946400
- PMCID: PMC8147151
- DOI: 10.3390/life11050412
DNA Methylation Change Profiling of Colorectal Disease: Screening towards Clinical Use
Abstract
Colon cancer remains one of the leading causes of cancer-related deaths worldwide. Transformation of colon epithelial cells into invasive adenocarcinomas has been well known to be due to the accumulation of multiple genetic and epigenetic changes. In the past decade, the etiology of inflammatory bowel disease (IBD) which is characterized by chronic inflammation of the intestinal mucosa, was only partially explained by genetic studies providing susceptibility loci, but recently epigenetic studies have provided critical evidences affecting IBD pathogenesis. Over the past decade, A deep understanding of epigenetics along with technological advances have led to identifying numerous genes that are regulated by promoter DNA hypermethylation in colorectal diseases. Recent advances in our understanding of the role of DNA methylation in colorectal diseases could improve a multitude of powerful DNA methylation-based biomarkers, particularly for use as diagnosis, prognosis, and prediction for therapeutic approaches. This review focuses on the emerging potential for translational research of epigenetic alterations into clinical utility as molecular biomarkers. Moreover, this review discusses recent progress regarding the identification of unknown hypermethylated genes in colon cancers and IBD, as well as their possible role in clinical practice, which will have important clinical significance, particularly in the era of the personalized medicine.
Keywords: DNA methylation; biomarkers; colorectal cancer; epigenetic regulation; inflammatory bowel diseases (IBDs).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
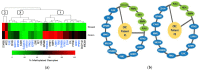


References
-
- Liang G., Lin J.C.Y., Wei V., Yoo C., Cheng J.C., Nguyen C.T., Weisenberger D.J., Egger G., Takai D., Gonzales F.A., et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. USA. 2004;101:7357. doi: 10.1073/pnas.0401866101. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

