Computational Identification of Potential Anti-Inflammatory Natural Compounds Targeting the p38 Mitogen-Activated Protein Kinase (MAPK): Implications for COVID-19-Induced Cytokine Storm
- PMID: 33946644
- PMCID: PMC8146027
- DOI: 10.3390/biom11050653
Computational Identification of Potential Anti-Inflammatory Natural Compounds Targeting the p38 Mitogen-Activated Protein Kinase (MAPK): Implications for COVID-19-Induced Cytokine Storm
Abstract
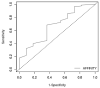
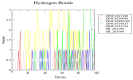
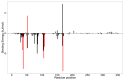
Severely ill coronavirus disease 2019 (COVID-19) patients show elevated concentrations of pro-inflammatory cytokines, a situation commonly known as a cytokine storm. The p38 MAPK receptor is considered a plausible therapeutic target because of its involvement in the platelet activation processes leading to inflammation. This study aimed to identify potential natural product-derived inhibitory molecules against the p38α MAPK receptor to mitigate the eliciting of pro-inflammatory cytokines using computational techniques. The 3D X-ray structure of the receptor with PDB ID 3ZS5 was energy minimized using GROMACS and used for molecular docking via AutoDock Vina. The molecular docking was validated with an acceptable area under the curve (AUC) of 0.704, which was computed from the receiver operating characteristic (ROC) curve. A compendium of 38,271 natural products originating from Africa and China together with eleven known p38 MAPK inhibitors were screened against the receptor. Four potential lead compounds ZINC1691180, ZINC5519433, ZINC4520996 and ZINC5733756 were identified. The compounds formed strong intermolecular bonds with critical residues Val38, Ala51, Lys53, Thr106, Leu108, Met109 and Phe169. Additionally, they exhibited appreciably low binding energies which were corroborated via molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) calculations. The compounds were also predicted to have plausible pharmacological profiles with insignificant toxicity. The molecules were also predicted to be anti-inflammatory, kinase inhibitors, antiviral, platelet aggregation inhibitors, and immunosuppressive, with probable activity (Pa) greater than probable inactivity (Pi). ZINC5733756 is structurally similar to estradiol with a Tanimoto coefficient value of 0.73, which exhibits anti-inflammatory activity by targeting the activation of Nrf2. Similarly, ZINC1691180 has been reported to elicit anti-inflammatory activity in vitro. The compounds may serve as scaffolds for the design of potential biotherapeutic molecules against the cytokine storm associated with COVID-19.
Keywords: COVID-19; anti-inflammatory compounds; coronavirus; cytokine storm; molecular docking; molecular dynamics simulation; natural products; p38 MAPK.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

