Emergence and Spread of SARS-CoV-2 Lineages B.1.1.7 and P.1 in Italy
- PMID: 33946747
- PMCID: PMC8146936
- DOI: 10.3390/v13050794
Emergence and Spread of SARS-CoV-2 Lineages B.1.1.7 and P.1 in Italy
Abstract
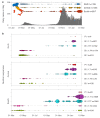
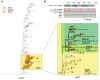
Italy's second wave of SARS-CoV-2 has hit hard, with more than three million cases and over 100,000 deaths, representing an almost ten-fold increase in the numbers reported by August 2020. Herein, we present an analysis of 6515 SARS-CoV-2 sequences sampled in Italy between 29 January 2020 and 1 March 2021 and show how different lineages emerged multiple times independently despite lockdown restrictions. Virus lineage B.1.177 became the dominant variant in November 2020, when cases peaked at 40,000 a day, but since January 2021 this is being replaced by the B.1.1.7 'variant of concern'. In addition, we report a sudden increase in another documented variant of concern-lineage P.1-from December 2020 onwards, most likely caused by a single introduction into Italy. We again highlight how international importations drive the emergence of new lineages and that genome sequencing should remain a top priority for ongoing surveillance in Italy.
Keywords: B.1.1.7; Italy; P.1; SARS-Cov-2; epidemic; variant of concern.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- WHO . Weekly Operational Update on COVID-19—8 March 2021. WHO; Geneva, Switzerland: 2021.
-
- WHO . Weekly Epidemiological Update—31 August 2020. WHO; Geneva, Switzerland: 2020.
-
- CDC Science Brief: Emerging SARS-CoV-2 Variants. [(accessed on 1 March 2021)]; Available online: https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scie....
-
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021 doi: 10.1126/science.abg3055. Online ahead of print. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

