Quo Vadis Caenorhabditis elegans Metabolomics-A Review of Current Methods and Applications to Explore Metabolism in the Nematode
- PMID: 33947148
- PMCID: PMC8146106
- DOI: 10.3390/metabo11050284
Quo Vadis Caenorhabditis elegans Metabolomics-A Review of Current Methods and Applications to Explore Metabolism in the Nematode
Abstract
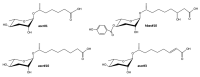
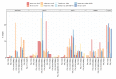
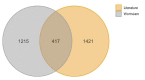
Metabolomics and lipidomics recently gained interest in the model organism Caenorhabditis elegans (C. elegans). The fast development, easy cultivation and existing forward and reverse genetic tools make the small nematode an ideal organism for metabolic investigations in development, aging, different disease models, infection, or toxicology research. The conducted type of analysis is strongly depending on the biological question and requires different analytical approaches. Metabolomic analyses in C. elegans have been performed using nuclear magnetic resonance (NMR) spectroscopy, direct infusion mass spectrometry (DI-MS), gas-chromatography mass spectrometry (GC-MS) and liquid chromatography mass spectrometry (LC-MS) or combinations of them. In this review we provide general information on the employed techniques and their advantages and disadvantages in regard to C. elegans metabolomics. Additionally, we reviewed different fields of application, e.g., longevity, starvation, aging, development or metabolism of secondary metabolites such as ascarosides or maradolipids. We also summarised applied bioinformatic tools that recently have been used for the evaluation of metabolomics or lipidomics data from C. elegans. Lastly, we curated metabolites and lipids from the reviewed literature, enabling a prototypic collection which serves as basis for a future C. elegans specific metabolome database.
Keywords: Caenorhabditis elegans; NMR; lipidomics; mass-spectrometry; metabolomics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

