Heterogenous Susceptibility to R-Pyocins in Populations of Pseudomonas aeruginosa Sourced from Cystic Fibrosis Lungs
- PMID: 33947755
- PMCID: PMC8262887
- DOI: 10.1128/mBio.00458-21
Heterogenous Susceptibility to R-Pyocins in Populations of Pseudomonas aeruginosa Sourced from Cystic Fibrosis Lungs
Abstract
Bacteriocins are proteinaceous antimicrobials produced by bacteria that are active against other strains of the same species. R-type pyocins are phage tail-like bacteriocins produced by Pseudomonas aeruginosa Due to their antipseudomonal activity, R-pyocins have potential as therapeutics in infection. P. aeruginosa is a Gram-negative opportunistic pathogen and is particularly problematic for individuals with cystic fibrosis (CF). P. aeruginosa organisms from CF lung infections develop increasing resistance to antibiotics, making new treatment approaches essential. P. aeruginosa populations become phenotypically and genotypically diverse during infection; however, little is known of the efficacy of R-pyocins against heterogeneous populations. R-pyocins vary by subtype (R1 to R5), distinguished by binding to different residues on the lipopolysaccharide (LPS). Each type varies in killing spectrum, and each strain produces only one R-type. To evaluate the prevalence of different R-types, we screened P. aeruginosa strains from the International Pseudomonas Consortium Database (IPCD) and from our biobank of CF strains. We found that (i) R1-types were the most prevalent R-type among strains from respiratory sources, (ii) a large number of strains lack R-pyocin genes, and (iii) isolates collected from the same patient have the same R-type. We then assessed the impact of intrastrain diversity on R-pyocin susceptibility and found a heterogenous response to R-pyocins within populations, likely due to differences in the LPS core. Our work reveals that heterogeneous populations of microbes exhibit variable susceptibility to R-pyocins and highlights that there is likely heterogeneity in response to other types of LPS-binding antimicrobials, including phage.IMPORTANCE R-pyocins have potential as alternative therapeutics against Pseudomonas aeruginosa in chronic infection; however, little is known about the efficacy of R-pyocins in heterogeneous bacterial populations. P. aeruginosa is known to become resistant to multiple antibiotics and to evolve phenotypic and genotypic diversity over time; thus, it is particularly difficult to eradicate in chronic cystic fibrosis (CF) lung infections. In this study, we found that P. aeruginosa populations from CF lungs maintain the same R-pyocin genotype but exhibit heterogeneity in susceptibility to R-pyocins from other strains. Our findings suggest there is heterogeneity in response to other types of LPS-binding antimicrobials, such as phage, highlighting the necessity of further studying the potential of LPS-binding antimicrobial particles as alternative therapies in chronic infections.
Keywords: Pseudomonas aeruginosa; R-pyocin; bacteriocin; bacteriocins; cystic fibrosis; heterogeneity; lipopolysaccharide; pyocins.
Copyright © 2021 Mei et al.
Figures


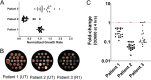

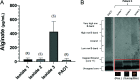

References
-
- Ito S, Kageyama M, Egami F. 1970. Isolation and characterization of pyocins from several strains of Pseudomonas aeruginosa. J Gen Appl Microbiol 16:205–214. doi:10.2323/jgam.16.3_205. - DOI
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

