Particulate Metabolites and Transcripts Reflect Diel Oscillations of Microbial Activity in the Surface Ocean
- PMID: 33947808
- PMCID: PMC8269247
- DOI: 10.1128/mSystems.00896-20
Particulate Metabolites and Transcripts Reflect Diel Oscillations of Microbial Activity in the Surface Ocean
Erratum in
-
Correction for Boysen et al., "Particulate Metabolites and Transcripts Reflect Diel Oscillations of Microbial Activity in the Surface Ocean".mSystems. 2023 Jun 29;8(3):e0108322. doi: 10.1128/msystems.01083-22. Epub 2023 Mar 21. mSystems. 2023. PMID: 36943103 Free PMC article. No abstract available.
Abstract
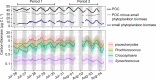
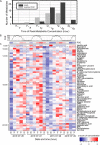
Light fuels photosynthesis and organic matter production by primary producers in the sunlit ocean. The quantity and quality of the organic matter produced influence community function, yet in situ measurements of metabolites, the products of cellular metabolism, over the diel cycle are lacking. We evaluated community-level biochemical consequences of oscillations of light in the North Pacific Subtropical Gyre by quantifying 79 metabolites in particulate organic matter from 15 m every 4 h over 8 days. Total particulate metabolite concentration peaked at dusk and represented up to 2% of total particulate organic carbon (POC). The concentrations of 55/79 (70%) individual metabolites exhibited significant 24-h periodicity, with daily fold changes from 1.6 to 12.8, often greater than those of POC and flow cytometry-resolvable biomass, which ranged from 1.2 to 2.8. Paired metatranscriptome analysis revealed the taxa involved in production and consumption of a subset of metabolites. Primary metabolites involved in anabolism and redox maintenance had significant 24-h periodicity and diverse organisms exhibited diel periodicity in transcript abundance associated with these metabolites. Compounds with osmotic properties displayed the largest oscillations in concentration, implying rapid turnover and supporting prior evidence of functions beyond cell turgor maintenance. The large daily oscillation of trehalose paired with metatranscriptome and culture data showed that trehalose is produced by the nitrogen-fixing cyanobacterium Crocosphaera, likely to store energy for nighttime metabolism. Together, paired measurements of particulate metabolites and transcripts resolve strategies that microbes use to manage daily energy and redox oscillations and highlight dynamic metabolites with cryptic roles in marine microbial ecosystems.IMPORTANCE Fueled by light, phytoplankton produce the organic matter that supports ocean ecosystems and carbon sequestration. Ocean change impacts microbial metabolism with repercussions for biogeochemical cycling. As the small molecule products of cellular metabolism, metabolites often change rapidly in response to environmental conditions and form the basis of energy and nutrient management and storage within cells. By pairing measurements of metabolites and gene expression in the stratified surface ocean, we reveal strategies of microbial energy management over the day-night cycle and hypothesize that oscillating metabolites are important substrates for dark respiration by phytoplankton. These high-resolution diel measurements of in situ metabolite concentrations form the basis for future work into the specific roles these compounds play in marine microbial communities.
Keywords: North Pacific; diel cycles; metabolomics; microbial ecology; oceanography; osmolytes; phytoplankton; transcriptomics.
Copyright © 2021 Boysen et al.
Figures




References
-
- Wakeham SG, Lee C, Hedges JI, Hernes PJ, Peterson MJ. 1997. Molecular indicators of diagenetic status in marine organic matter. Geochim Cosmochim Acta 61:5363–5369. doi:10.1016/S0016-7037(97)00312-8. - DOI
-
- Hedges JI, Baldock JA, Gélinas Y, Lee C, Peterson ML, Wakeham SG. 2002. The biochemical and elemental compositions of marine plankton: a NMR perspective. Mar Chem 78:47–63. doi:10.1016/S0304-4203(02)00009-9. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

