Differential chromatin binding of the lung lineage transcription factor NKX2-1 resolves opposing murine alveolar cell fates in vivo
- PMID: 33947861
- PMCID: PMC8096971
- DOI: 10.1038/s41467-021-22817-6
Differential chromatin binding of the lung lineage transcription factor NKX2-1 resolves opposing murine alveolar cell fates in vivo
Abstract
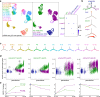
Differential transcription of identical DNA sequences leads to distinct tissue lineages and then multiple cell types within a lineage, an epigenetic process central to progenitor and stem cell biology. The associated genome-wide changes, especially in native tissues, remain insufficiently understood, and are hereby addressed in the mouse lung, where the same lineage transcription factor NKX2-1 promotes the diametrically opposed alveolar type 1 (AT1) and AT2 cell fates. Here, we report that the cell-type-specific function of NKX2-1 is attributed to its differential chromatin binding that is acquired or retained during development in coordination with partner transcriptional factors. Loss of YAP/TAZ redirects NKX2-1 from its AT1-specific to AT2-specific binding sites, leading to transcriptionally exaggerated AT2 cells when deleted in progenitors or AT1-to-AT2 conversion when deleted after fate commitment. Nkx2-1 mutant AT1 and AT2 cells gain distinct chromatin accessible sites, including those specific to the opposite fate while adopting a gastrointestinal fate, suggesting an epigenetic plasticity unexpected from transcriptional changes. Our genomic analysis of single or purified cells, coupled with precision genetics, provides an epigenetic basis for alveolar cell fate and potential, and introduces an experimental benchmark for deciphering the in vivo function of lineage transcription factors.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

