Serial profiling of cell-free DNA and nucleosome histone modifications in cell cultures
- PMID: 33947882
- PMCID: PMC8096822
- DOI: 10.1038/s41598-021-88866-5
Serial profiling of cell-free DNA and nucleosome histone modifications in cell cultures
Abstract
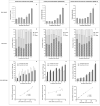
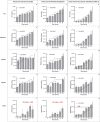
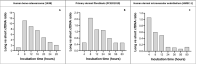
Recent advances in basic research have unveiled several strategies for improving the sensitivity and specificity of cell-free DNA (cfDNA) based assays, which is a prerequisite for broadening its clinical use. Included among these strategies is leveraging knowledge of both the biogenesis and physico-chemical properties of cfDNA towards the identification of better disease-defining features and optimization of methods. While good progress has been made on this front, much of cfDNA biology remains uncharted. Here, we correlated serial measurements of cfDNA size, concentration and nucleosome histone modifications with various cellular parameters, including cell growth rate, viability, apoptosis, necrosis, and cell cycle phase in three different cell lines. Collectively, the picture emerged that temporal changes in cfDNA levels are rather irregular and not the result of constitutive release from live cells. Instead, changes in cfDNA levels correlated with intermittent cell death events, wherein apoptosis contributed more to cfDNA release in non-cancer cells and necrosis more in cancer cells. Interestingly, the presence of a ~ 3 kbp cfDNA population, which is often deemed to originate from accidental cell lysis or active release, was found to originate from necrosis. High-resolution analysis of this cfDNA population revealed an underlying DNA laddering pattern consisting of several oligo-nucleosomes, identical to those generated by apoptosis. This suggests that necrosis may contribute significantly to the pool of mono-nucleosomal cfDNA fragments that are generally interrogated for cancer mutational profiling. Furthermore, since active steps are often taken to exclude longer oligo-nucleosomes from clinical biospecimens and subsequent assays this raises the question of whether important pathological information is lost.
Conflict of interest statement
M.H. and P.vd.A. are employed by Belgian Volition SRL (Namur, Belgium). S.H. serves on the Scientific Advisory Board and acts as a consultant for Belgian Volition SRL (Namur, Belgium). V.U. and A.J.B. declare no competing interests.
Figures




References
-
- Heitzer, E., Haque, I. S., Roberts, C. E. S. & Speicher, M. R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 1–1 (2018). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

