Systematic integrated analyses of methylomic and transcriptomic impacts of early combined botanicals on estrogen receptor-negative mammary cancer
- PMID: 33947955
- PMCID: PMC8096837
- DOI: 10.1038/s41598-021-89131-5
Systematic integrated analyses of methylomic and transcriptomic impacts of early combined botanicals on estrogen receptor-negative mammary cancer
Abstract
Dietary botanicals such as the cruciferous vegetable broccoli sprouts (BSp) as well as green tea polyphenols (GTPs) have shown exciting potential in preventing or delaying breast cancer (BC). However, little is known about their impact on epigenomic aberrations that are centrally involved in the initiation and progression of estrogen receptor-negative [ER(-)] BC. We have investigated the efficacy of combined BSp and GTPs diets on mammary tumor inhibition in transgenic Her2/neu mice that were administered the diets from prepubescence until adulthood. Herein, we present an integrated DNA methylome and transcriptome analyses for defining the early-life epigenetic impacts of combined BSp and GTPs on mammary tumors and our results indicate that a combinatorial administration of BSp and GTPs have a stronger impact at both transcriptome and methylome levels in comparison to BSp or GTPs administered alone. We also demonstrated a streamlined approach by performing an extensive preprocessing, quality assessment and downstream analyses on the genomic dataset. Our identification of differentially methylated regions in response to dietary botanicals administered during early-life will allow us to identify key genes and facilitate implementation of the subsequent downstream functional analyses on a genomic scale and various epigenetic modifications that are crucial in preventing ER(-) mammary cancer. Furthermore, our realtime PCR results were also found to be consistent with our genome-wide analysis results. These results could be exploited as a comprehensive resource for understanding understudied genes and their associated epigenetic modifications in response to these dietary botanicals.
Conflict of interest statement
The authors declare no competing interests.
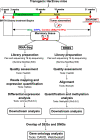
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

