A highly multiplexed droplet digital PCR assay to measure the intact HIV-1 proviral reservoir
- PMID: 33948574
- PMCID: PMC8080125
- DOI: 10.1016/j.xcrm.2021.100243
A highly multiplexed droplet digital PCR assay to measure the intact HIV-1 proviral reservoir
Abstract
Quantifying the replication-competent HIV reservoir is essential for evaluating curative strategies. Viral outgrowth assays (VOAs) underestimate the reservoir because they fail to induce all replication-competent proviruses. Single- or double-region HIV DNA assays overestimate it because they fail to exclude many defective proviruses. We designed two triplex droplet digital PCR assays, each with 2 unique targets and 1 in common, and normalize the results to PCR-based T cell counts. Both HIV assays are specific, sensitive, and reproducible. Together, they estimate the number of proviruses containing all five primer-probe regions. Our 5-target results are on average 12.1-fold higher than and correlate with paired quantitative VOA (Spearman's ρ = 0.48) but estimate a markedly smaller reservoir than previous DNA assays. In patients on antiretroviral therapy, decay rates in blood CD4+ T cells are faster for intact than for defective proviruses, and intact provirus frequencies are similar in mucosal and circulating T cells.
Keywords: HIV cure; HIV reservoir; IPDA; digital PCR; genital mucosa; intact proviral DNA assay; intestinal mucosa; multiplexing; rectal; viral outgrowth assay.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

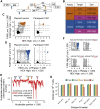

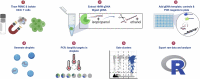



Comment in
-
Measuring the size and decay dynamics of the HIV-1 latent reservoir.Cell Rep Med. 2021 Apr 20;2(4):100249. doi: 10.1016/j.xcrm.2021.100249. eCollection 2021 Apr 20. Cell Rep Med. 2021. PMID: 33948579 Free PMC article.
References
-
- Wonderlich E.R., Subramanian K., Cox B., Wiegand A., Lackman-Smith C., Bale M.J., Stone M., Hoh R., Kearney M.F., Maldarelli F. Effector memory differentiation increases detection of replication-competent HIV-l in resting CD4+ T cells from virally suppressed individuals. PLoS Pathog. 2019;15:e1008074. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI071212/AI/NIAID NIH HHS/United States
- K25 AI155224/AI/NIAID NIH HHS/United States
- T32 AI007140/AI/NIAID NIH HHS/United States
- R01 DA040386/DA/NIDA NIH HHS/United States
- R01 AI116292/AI/NIAID NIH HHS/United States
- UM1 AI126611/AI/NIAID NIH HHS/United States
- UM1 AI126623/AI/NIAID NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- P30 AI064518/AI/NIAID NIH HHS/United States
- R01 AI125026/AI/NIAID NIH HHS/United States
- HHSN272201500017C/AI/NIAID NIH HHS/United States
- R01 HD094718/HD/NICHD NIH HHS/United States
- P30 AI027757/AI/NIAID NIH HHS/United States
- UM1 AI106701/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

