Probing the oligomeric re-assembling of bacterial fimbriae in vitro: a small-angle X-ray scattering and analytical ultracentrifugation study
- PMID: 33948690
- PMCID: PMC8190007
- DOI: 10.1007/s00249-021-01543-3
Probing the oligomeric re-assembling of bacterial fimbriae in vitro: a small-angle X-ray scattering and analytical ultracentrifugation study
Abstract
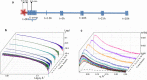
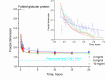
Capsular antigen fragment 1 (Caf1) is an oligomeric protein consisting of 15 kDa monomeric subunits that are non-covalently linked through exceptionally strong and kinetically inert interactions into a linear polymer chain. It has been shown that after its thermal depolymerisation into unfolded monomeric subunits, Caf1 is able to efficiently repolymerise in vitro to reform its polymeric structure. However, little is known about the nature of the repolymerisation process. An improved understanding of this process will lead to the development of methods to better control the lengths of the repolymerised species, and ultimately, to better design of the properties of Caf1-based materials. Here we utilize small-angle X-ray scattering to estimate the size of Caf1 polymers during the first 24 h of the re-polymerisation process. Analytical ultracentrifugation measurements were also used to investigate the process post-24 h, where the rate of repolymerisation becomes considerably slower. Results show that in vitro polymerisation proceeds in a linear manner with no evidence observed for the formation of a lateral polymer network or uncontrolled aggregates. The rate of Caf1 in vitro repolymerisation was found to be concentration-dependent. Importantly, the rate of polymer growth was found to be relatively fast over the first few hours, before continuing at a dramatically slower rate. This observation is not consistent with the previously proposed step-growth mechanism of in vitro polymerisation of Caf1, where a linear increase in polymer length would be expected with time. We speculate how our observations may support the idea that the polymerisation process may be occurring at the ends of the chains with monomers adding sequentially. Our findings will contribute towards the development of new biomaterials for 3D cell culture and bio-printing.
Keywords: Flexibility; Mass increment; Oligomeric growth; Repolymerisation.
Figures





Similar articles
-
The polymer and materials science of the bacterial fimbriae Caf1.Biomater Sci. 2023 Nov 7;11(22):7229-7246. doi: 10.1039/d3bm01075a. Biomater Sci. 2023. PMID: 37791425 Free PMC article. Review.
-
Hydrogels of engineered bacterial fimbriae can finely tune 2D human cell culture.Biomater Sci. 2021 Apr 7;9(7):2542-2552. doi: 10.1039/d0bm01966f. Epub 2021 Feb 11. Biomater Sci. 2021. PMID: 33571331
-
Engineered mosaic protein polymers; a simple route to multifunctional biomaterials.J Biol Eng. 2019 Jun 18;13:54. doi: 10.1186/s13036-019-0183-2. eCollection 2019. J Biol Eng. 2019. PMID: 31244892 Free PMC article.
-
Large is fast, small is tight: determinants of speed and affinity in subunit capture by a periplasmic chaperone.J Mol Biol. 2012 Apr 6;417(4):294-308. doi: 10.1016/j.jmb.2012.01.020. Epub 2012 Feb 1. J Mol Biol. 2012. PMID: 22321795
-
Quaternary Structure Analyses of an Essential Oligomeric Enzyme.Methods Enzymol. 2015;562:205-23. doi: 10.1016/bs.mie.2015.06.020. Epub 2015 Aug 13. Methods Enzymol. 2015. PMID: 26412653 Review.
Cited by
-
Protein quaternary structures in solution are a mixture of multiple forms.Chem Sci. 2022 Sep 21;13(39):11680-11695. doi: 10.1039/d2sc02794a. eCollection 2022 Oct 12. Chem Sci. 2022. PMID: 36320402 Free PMC article.
-
Community-building and promotion of technological excellence in molecular biophysics: the ARBRE-MOBIEU network.Eur Biophys J. 2021 May;50(3-4):307-311. doi: 10.1007/s00249-021-01550-4. Eur Biophys J. 2021. PMID: 34057541 No abstract available.
-
The polymer and materials science of the bacterial fimbriae Caf1.Biomater Sci. 2023 Nov 7;11(22):7229-7246. doi: 10.1039/d3bm01075a. Biomater Sci. 2023. PMID: 37791425 Free PMC article. Review.
References
-
- Bernado P. Effect of interdomain dynamics on the structure determination of modular proteins by small-angle scattering. Eur Biophys J. 2010;39:769–780. - PubMed
-
- Bernadó P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. - PubMed
-
- Brookes E, Demeler B, Rocco M. Developments in the US-SOMO bead modeling suite: new features in the direct residue-to-bead method, improved grid routines, and influence of accessible surface area screening. Macromol Biosci. 2010;10:746–753. - PubMed
-
- Brookes E, Demeler B, Rosano C, Rocco M. The implementation of SOMO (SOlution MOdeller) in the UltraScan analytical ultracentrifugation data analysis suite: enhanced capabilities allow the reliable hydrodynamic modeling of virtually any kind of biomacromolecule. Eur Biophys J. 2010;39:423–435. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

