Proteomic analysis identifies key differences in the cardiac interactomes of dystrophin and micro-dystrophin
- PMID: 33949649
- PMCID: PMC8255133
- DOI: 10.1093/hmg/ddab133
Proteomic analysis identifies key differences in the cardiac interactomes of dystrophin and micro-dystrophin
Abstract
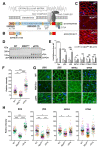
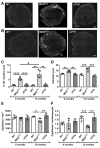
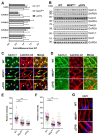
ΔR4-R23/ΔCT micro-dystrophin (μDys) is a miniaturized version of dystrophin currently evaluated in a Duchenne muscular dystrophy (DMD) gene therapy trial to treat skeletal and cardiac muscle disease. In pre-clinical studies, μDys efficiently rescues cardiac histopathology, but only partially normalizes cardiac function. To gain insights into factors that may impact the cardiac therapeutic efficacy of μDys, we compared by mass spectrometry the composition of purified dystrophin and μDys protein complexes in the mouse heart. We report that compared to dystrophin, μDys has altered associations with α1- and β2-syntrophins, as well as cavins, a group of caveolae-associated signaling proteins. In particular, we found that membrane localization of cavin-1 and cavin-4 in cardiomyocytes requires dystrophin and is profoundly disrupted in the heart of mdx5cv mice, a model of DMD. Following cardiac stress/damage, membrane-associated cavin-4 recruits the signaling molecule ERK to caveolae, which activates key cardio-protective responses. Evaluation of ERK signaling revealed a profound inhibition, below physiological baseline, in the mdx5cv mouse heart. Expression of μDys in mdx5cv mice prevented the development of cardiac histopathology but did not rescue membrane localization of cavins nor did it normalize ERK signaling. Our study provides the first comparative analysis of purified protein complexes assembled in vivo by full-length dystrophin and a therapeutic micro-dystrophin construct. This has revealed disruptions in cavins and ERK signaling that may contribute to DMD cardiomyopathy. This new knowledge is important for ongoing efforts to prevent and treat heart disease in DMD patients.
© The Author(s) 2021. Published by Oxford University Press.
Figures


Similar articles
-
AAV micro-dystrophin gene therapy alleviates stress-induced cardiac death but not myocardial fibrosis in >21-m-old mdx mice, an end-stage model of Duchenne muscular dystrophy cardiomyopathy.J Mol Cell Cardiol. 2012 Aug;53(2):217-22. doi: 10.1016/j.yjmcc.2012.05.002. Epub 2012 May 12. J Mol Cell Cardiol. 2012. PMID: 22587991 Free PMC article.
-
Development of Novel Micro-dystrophins with Enhanced Functionality.Mol Ther. 2019 Mar 6;27(3):623-635. doi: 10.1016/j.ymthe.2019.01.002. Epub 2019 Feb 1. Mol Ther. 2019. PMID: 30718090 Free PMC article.
-
Injection of vessel-derived stem cells prevents dilated cardiomyopathy and promotes angiogenesis and endogenous cardiac stem cell proliferation in mdx/utrn-/- but not aged mdx mouse models for duchenne muscular dystrophy.Stem Cells Transl Med. 2013 Jan;2(1):68-80. doi: 10.5966/sctm.2012-0107. Epub 2012 Dec 27. Stem Cells Transl Med. 2013. PMID: 23283493 Free PMC article.
-
Proteomic profiling of the dystrophin-deficient mdx phenocopy of dystrophinopathy-associated cardiomyopathy.Biomed Res Int. 2014;2014:246195. doi: 10.1155/2014/246195. Epub 2014 Mar 20. Biomed Res Int. 2014. PMID: 24772416 Free PMC article. Review.
-
Brain function in Duchenne muscular dystrophy.Brain. 2002 Jan;125(Pt 1):4-13. doi: 10.1093/brain/awf012. Brain. 2002. PMID: 11834588 Review.
Cited by
-
The 2022 On-site Padua Days on Muscle and Mobility Medicine hosts the University of Florida Institute of Myology and the Wellstone Center, March 30 - April 3, 2022 at the University of Padua and Thermae of Euganean Hills, Padua, Italy: The collection of abstracts.Eur J Transl Myol. 2022 Mar 10;32(1):10440. doi: 10.4081/ejtm.2022.10440. Eur J Transl Myol. 2022. PMID: 35272451 Free PMC article.
-
Therapeutic Strategies for Dystrophin Replacement in Duchenne Muscular Dystrophy.Front Med (Lausanne). 2022 Mar 28;9:859930. doi: 10.3389/fmed.2022.859930. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35419381 Free PMC article. Review.
-
Systemic delivery of full-length dystrophin in Duchenne muscular dystrophy mice.Nat Commun. 2024 Jul 21;15(1):6141. doi: 10.1038/s41467-024-50569-6. Nat Commun. 2024. PMID: 39034316 Free PMC article.
-
Lamin A precursor localizes to the Z-disc of sarcomeres in the heart and is dynamically regulated in muscle cell differentiation.Philos Trans R Soc Lond B Biol Sci. 2022 Nov 21;377(1864):20210490. doi: 10.1098/rstb.2021.0490. Epub 2022 Oct 3. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 36189817 Free PMC article.
-
The role of the dystrophin glycoprotein complex in muscle cell mechanotransduction.Commun Biol. 2022 Sep 27;5(1):1022. doi: 10.1038/s42003-022-03980-y. Commun Biol. 2022. PMID: 36168044 Free PMC article. Review.
References
-
- Hor, K.N., Mah, M.L., Johnston, P., Cripe, T.P. and Cripe, L.H. (2018) Advances in the diagnosis and management of cardiomyopathy in Duchenne muscular dystrophy. Neuromuscul. Disord., 28, 711–716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

