SARS-CoV-2 RNAemia Predicts Clinical Deterioration and Extrapulmonary Complications from COVID-19
- PMID: 33949665
- PMCID: PMC8135992
- DOI: 10.1093/cid/ciab394
SARS-CoV-2 RNAemia Predicts Clinical Deterioration and Extrapulmonary Complications from COVID-19
Abstract
Background: The determinants of coronavirus disease 2019 (COVID-19) disease severity and extrapulmonary complications (EPCs) are poorly understood. We characterized relationships between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNAemia and disease severity, clinical deterioration, and specific EPCs.
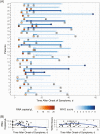
Methods: We used quantitative and digital polymerase chain reaction (qPCR and dPCR) to quantify SARS-CoV-2 RNA from plasma in 191 patients presenting to the emergency department with COVID-19. We recorded patient symptoms, laboratory markers, and clinical outcomes, with a focus on oxygen requirements over time. We collected longitudinal plasma samples from a subset of patients. We characterized the role of RNAemia in predicting clinical severity and EPCs using elastic net regression.
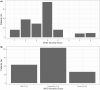
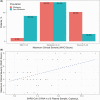
Results: Of SARS-CoV-2-positive patients, 23.0% (44 of 191) had viral RNA detected in plasma by dPCR, compared with 1.4% (2 of 147) by qPCR. Most patients with serial measurements had undetectable RNAemia within 10 days of symptom onset, reached maximum clinical severity within 16 days, and symptom resolution within 33 days. Initially RNAemic patients were more likely to manifest severe disease (odds ratio, 6.72 [95% confidence interval, 2.45-19.79]), worsening of disease severity (2.43 [1.07-5.38]), and EPCs (2.81 [1.26-6.36]). RNA loads were correlated with maximum severity (r = 0.47 [95% confidence interval, .20-.67]).
Conclusions: dPCR is more sensitive than qPCR for the detection of SARS-CoV-2 RNAemia, which is a robust predictor of eventual COVID-19 severity and oxygen requirements, as well as EPCs. Because many COVID-19 therapies are initiated on the basis of oxygen requirements, RNAemia on presentation might serve to direct early initiation of appropriate therapies for the patients most likely to deteriorate.
Keywords: RNAemia; SARS-CoV-2; digital PCR; extrapulmonary complications; severity prediction.
© The Author(s) 2021. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures

Update of
-
SARS-CoV-2 RNAaemia predicts clinical deterioration and extrapulmonary complications from COVID-19.medRxiv [Preprint]. 2020 Dec 22:2020.12.19.20248561. doi: 10.1101/2020.12.19.20248561. medRxiv. 2020. Update in: Clin Infect Dis. 2022 Jan 29;74(2):218-226. doi: 10.1093/cid/ciab394. PMID: 33398290 Free PMC article. Updated. Preprint.
References
-
- WHO coronavirus (COVID-19) dashboard. Available at: https://covid19.who.int. Accessed 18 April 2021.
-
- Peng L, Liu J, Xu W, et al. 2019 Novel coronavirus can be detected in urine, blood, anal swabs and oropharyngeal swabs samples. medRxiv [Preprint: not peer reviewed]. 25 February 2020. Available from: https://www.medrxiv.org/content/10.1101/2020.02.21.20026179v1. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

