Ribosome dynamics and mRNA turnover, a complex relationship under constant cellular scrutiny
- PMID: 33949788
- PMCID: PMC8519046
- DOI: 10.1002/wrna.1658
Ribosome dynamics and mRNA turnover, a complex relationship under constant cellular scrutiny
Abstract
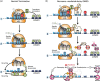
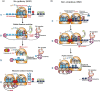
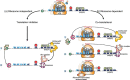
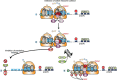
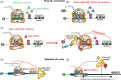
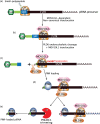
Eukaryotic gene expression is closely regulated by translation and turnover of mRNAs. Recent advances highlight the importance of translation in the control of mRNA degradation, both for aberrant and apparently normal mRNAs. During translation, the information contained in mRNAs is decoded by ribosomes, one codon at a time, and tRNAs, by specifically recognizing codons, translate the nucleotide code into amino acids. Such a decoding step does not process regularly, with various obstacles that can hinder ribosome progression, then leading to ribosome stalling or collisions. The progression of ribosomes is constantly monitored by the cell which has evolved several translation-dependent mRNA surveillance pathways, including nonsense-mediated decay (NMD), no-go decay (NGD), and non-stop decay (NSD), to degrade certain problematic mRNAs and the incomplete protein products. Recent progress in sequencing and ribosome profiling has made it possible to discover new mechanisms controlling ribosome dynamics, with numerous crosstalks between translation and mRNA decay. We discuss here various translation features critical for mRNA decay, with particular focus on current insights from the complexity of the genetic code and also the emerging role for the ribosome as a regulatory hub orchestrating mRNA decay, quality control, and stress signaling. Even if the interplay between mRNA translation and degradation is no longer to be demonstrated, a better understanding of their precise coordination is worthy of further investigation. This article is categorized under: RNA Turnover and Surveillance > Regulation of RNA Stability Translation > Translation Regulation RNA Interactions with Proteins and Other Molecules > RNA-Protein Complexes.
Keywords: decay; degradation; mRNA; ribosome; translation.
© 2021 The Authors. WIREs RNA published by Wiley Periodicals LLC.
Conflict of interest statement
The authors have declared no conflicts of interest for this article.
Figures

Similar articles
-
A comprehensive coverage insurance for cells: revealing links between ribosome collisions, stress responses and mRNA surveillance.RNA Biol. 2022;19(1):609-621. doi: 10.1080/15476286.2022.2065116. Epub 2021 Dec 31. RNA Biol. 2022. PMID: 35491909 Free PMC article. Review.
-
New insights into no-go, non-stop and nonsense-mediated mRNA decay complexes.Curr Opin Struct Biol. 2020 Dec;65:110-118. doi: 10.1016/j.sbi.2020.06.011. Epub 2020 Jul 17. Curr Opin Struct Biol. 2020. PMID: 32688260 Review.
-
Features and factors that dictate if terminating ribosomes cause or counteract nonsense-mediated mRNA decay.J Biol Chem. 2022 Nov;298(11):102592. doi: 10.1016/j.jbc.2022.102592. Epub 2022 Oct 13. J Biol Chem. 2022. PMID: 36244451 Free PMC article. Review.
-
Human NMD ensues independently of stable ribosome stalling.Nat Commun. 2020 Aug 17;11(1):4134. doi: 10.1038/s41467-020-17974-z. Nat Commun. 2020. PMID: 32807779 Free PMC article.
-
Ribosome-based quality control of mRNA and nascent peptides.Wiley Interdiscip Rev RNA. 2017 Jan;8(1):10.1002/wrna.1366. doi: 10.1002/wrna.1366. Epub 2016 May 18. Wiley Interdiscip Rev RNA. 2017. PMID: 27193249 Free PMC article. Review.
Cited by
-
A rapid inducible RNA decay system reveals fast mRNA decay in P-bodies.Nat Commun. 2024 Mar 28;15(1):2720. doi: 10.1038/s41467-024-46943-z. Nat Commun. 2024. PMID: 38548718 Free PMC article.
-
Ribosome stalling-induced NIP5;1 mRNA decay triggers ARGONAUTE1-dependent transcription downregulation.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf159. doi: 10.1093/nar/gkaf159. Nucleic Acids Res. 2025. PMID: 40107731 Free PMC article.
-
Ribosome Fate during Decoding of UGA-Sec Codons.Int J Mol Sci. 2021 Dec 8;22(24):13204. doi: 10.3390/ijms222413204. Int J Mol Sci. 2021. PMID: 34948001 Free PMC article. Review.
-
The role of molecular chaperone CCT/TRiC in translation elongation: A literature review.Heliyon. 2024 Apr 1;10(7):e29029. doi: 10.1016/j.heliyon.2024.e29029. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38596045 Free PMC article. Review.
-
Newly synthesized mRNA escapes translational repression during the acute phase of the mammalian unfolded protein response.PLoS One. 2022 Aug 10;17(8):e0271695. doi: 10.1371/journal.pone.0271695. eCollection 2022. PLoS One. 2022. PMID: 35947624 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

