CM1-driven assembly and activation of yeast γ-tubulin small complex underlies microtubule nucleation
- PMID: 33949948
- PMCID: PMC8099430
- DOI: 10.7554/eLife.65168
CM1-driven assembly and activation of yeast γ-tubulin small complex underlies microtubule nucleation
Abstract
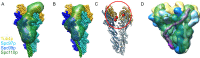
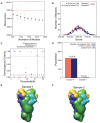
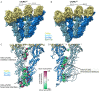
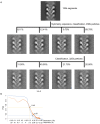
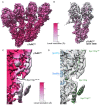
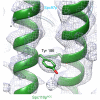
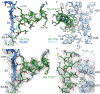
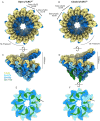
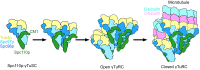
Microtubule (MT) nucleation is regulated by the γ-tubulin ring complex (γTuRC), conserved from yeast to humans. In Saccharomyces cerevisiae, γTuRC is composed of seven identical γ-tubulin small complex (γTuSC) sub-assemblies, which associate helically to template MT growth. γTuRC assembly provides a key point of regulation for the MT cytoskeleton. Here, we combine crosslinking mass spectrometry, X-ray crystallography, and cryo-EM structures of both monomeric and dimeric γTuSCs, and open and closed helical γTuRC assemblies in complex with Spc110p to elucidate the mechanisms of γTuRC assembly. γTuRC assembly is substantially aided by the evolutionarily conserved CM1 motif in Spc110p spanning a pair of adjacent γTuSCs. By providing the highest resolution and most complete views of any γTuSC assembly, our structures allow phosphorylation sites to be mapped, surprisingly suggesting that they are mostly inhibitory. A comparison of our structures with the CM1 binding site in the human γTuRC structure at the interface between GCP2 and GCP6 allows for the interpretation of significant structural changes arising from CM1 helix binding to metazoan γTuRC.
Keywords: S. cerevisiae; cell biology; cytoskeleton; electron microscopy; microtubule nucleation; molecular biophysics; structural biology.
© 2021, Brilot et al.
Conflict of interest statement
AB, AL, AZ, SV, AM, MM, EM, AS, TD, DA No competing interests declared
Figures















References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX : a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with ıt phenix.refine. Acta Crystallographica Section D. 2012;68:352–367. doi: 10.1107/S0907444912001308. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 GM105537/GM/NIGMS NIH HHS/United States
- 0714/HHMI/Howard Hughes Medical Institute/United States
- P30 GM124169/GM/NIGMS NIH HHS/United States
- R35 GM118099/GM/NIGMS NIH HHS/United States
- R01 GM124149/GM/NIGMS NIH HHS/United States
- P41 GM103533/GM/NIGMS NIH HHS/United States
- S10 OD021741/OD/NIH HHS/United States
- R01 GM083960/GM/NIGMS NIH HHS/United States
- R01 GM104556/GM/NIGMS NIH HHS/United States
- R01 GM031627/GM/NIGMS NIH HHS/United States
- S10 OD020054/OD/NIH HHS/United States
- R35 GM130293/GM/NIGMS NIH HHS/United States
- P41 GM109824/GM/NIGMS NIH HHS/United States
- S10 OD021596/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

