Extracting Drug Names and Associated Attributes From Discharge Summaries: Text Mining Study
- PMID: 33949962
- PMCID: PMC8135022
- DOI: 10.2196/24678
Extracting Drug Names and Associated Attributes From Discharge Summaries: Text Mining Study
Abstract
Background: Drug prescriptions are often recorded in free-text clinical narratives; making this information available in a structured form is important to support many health-related tasks. Although several natural language processing (NLP) methods have been proposed to extract such information, many challenges remain.
Objective: This study evaluates the feasibility of using NLP and deep learning approaches for extracting and linking drug names and associated attributes identified in clinical free-text notes and presents an extensive error analysis of different methods. This study initiated with the participation in the 2018 National NLP Clinical Challenges (n2c2) shared task on adverse drug events and medication extraction.
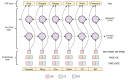
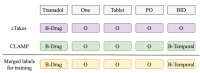
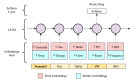
Methods: The proposed system (DrugEx) consists of a named entity recognizer (NER) to identify drugs and associated attributes and a relation extraction (RE) method to identify the relations between them. For NER, we explored deep learning-based approaches (ie, bidirectional long-short term memory with conditional random fields [BiLSTM-CRFs]) with various embeddings (ie, word embedding, character embedding [CE], and semantic-feature embedding) to investigate how different embeddings influence the performance. A rule-based method was implemented for RE and compared with a context-aware long-short term memory (LSTM) model. The methods were trained and evaluated using the 2018 n2c2 shared task data.
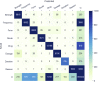
Results: The experiments showed that the best model (BiLSTM-CRFs with pretrained word embeddings [PWE] and CE) achieved lenient micro F-scores of 0.921 for NER, 0.927 for RE, and 0.855 for the end-to-end system. NER, which relies on the pretrained word and semantic embeddings, performed better on most individual entity types, but NER with PWE and CE had the highest classification efficiency among the proposed approaches. Extracting relations using the rule-based method achieved higher accuracy than the context-aware LSTM for most relations. Interestingly, the LSTM model performed notably better in the reason-drug relations, the most challenging relation type.
Conclusions: The proposed end-to-end system achieved encouraging results and demonstrated the feasibility of using deep learning methods to extract medication information from free-text data.
Keywords: discharge summaries; electronic health records; information extraction; medication prescriptions; natural language processing.
©Ghada Alfattni, Maksim Belousov, Niels Peek, Goran Nenadic. Originally published in JMIR Medical Informatics (https://medinform.jmir.org), 05.05.2021.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures

References
-
- Abhyankar S, Demner-Fushman D, Callaghan FM, McDonald CJ. Combining structured and unstructured data to identify a cohort of ICU patients who received dialysis. J Am Med Inform Assoc. 2014;21(5):801–7. doi: 10.1136/amiajnl-2013-001915. http://europepmc.org/abstract/MED/24384230 - DOI - PMC - PubMed
-
- Evans DA, Brownlow ND, Hersh WR, Campbell EM. Automating concept identification in the electronic medical record: an experiment in extracting dosage information. Proc AMIA Annu Fall Symp. 1996:388–92. http://europepmc.org/abstract/MED/8947694 - PMC - PubMed
-
- Karystianis G. Extraction and representation of key characteristics from epidemiological literature. The University of Manchester. 2014. [2021-03-31]. https://tinyurl.com/bv927sfthttps://tinyurl.com/645sksnd.
-
- MacKinlay AD, Verspoor KM. Extracting structured information from free-text medication prescriptions using dependencies. Proceedings of the ACM sixth international workshop on Data and text mining in biomedical informatics; CIKM'12: 21st ACM International Conference on Information and Knowledge Management; October, 2012; Maui Hawaii USA. 2012. pp. 35–40. - DOI
-
- Sohn S, Clark C, Halgrim SR, Murphy SP, Chute CG, Liu H. MedXN: an open source medication extraction and normalization tool for clinical text. J Am Med Inform Assoc. 2014;21(5):858–65. doi: 10.1136/amiajnl-2013-002190. http://europepmc.org/abstract/MED/24637954 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

