Genomic patterns of structural variation among diverse genotypes of Sorghum bicolor and a potential role for deletions in local adaptation
- PMID: 33950177
- PMCID: PMC8495935
- DOI: 10.1093/g3journal/jkab154
Genomic patterns of structural variation among diverse genotypes of Sorghum bicolor and a potential role for deletions in local adaptation
Abstract
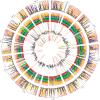
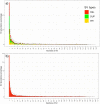
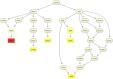
Genomic structural mutations, especially deletions, are an important source of variation in many species and can play key roles in phenotypic diversification and evolution. Previous work in many plant species has identified multiple instances of structural variations (SVs) occurring in or near genes related to stress response and disease resistance, suggesting a possible role for SVs in local adaptation. Sorghum [Sorghum bicolor (L.) Moench] is one of the most widely grown cereal crops in the world. It has been adapted to an array of different climates as well as bred for multiple purposes, resulting in a striking phenotypic diversity. In this study, we identified genome-wide SVs in the Biomass Association Panel, a collection of 347 diverse sorghum genotypes collected from multiple countries and continents. Using Illumina-based, short-read whole-genome resequencing data from every genotype, we found a total of 24,648 SVs, including 22,359 deletions. The global site frequency spectrum of deletions and other types of SVs fit a model of neutral evolution, suggesting that the majority of these mutations were not under any types of selection. Clustering results based on single nucleotide polymorphisms separated the genotypes into eight clusters which largely corresponded with geographic origins, with many of the large deletions we uncovered being unique to a single cluster. Even though most deletions appeared to be neutral, a handful of cluster-specific deletions were found in genes related to biotic and abiotic stress responses, supporting the possibility that at least some of these deletions contribute to local adaptation in sorghum.
Keywords: Sorghum; k-mean clustering; local adaptation; structural variation.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures



References
-
- Alexa A, Rahnenfuhrer J.. 2020. topGO: Enrichment Analysis for Gene Ontology. R package version 2.42.0. Rahnenfuhrer.
-
- Alexander DH, Shringarpure SS, Novembre J, Lange K.. 2015. Admixture 1.3 software manual. Los Angeles, CA: UCLA Human Genetics Software Distribution.
-
- Banerjee A, Roychoudhury A.. 2017. Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma. 254:3–16. - PubMed
-
- Barton KE, Jones C, Edwards KF, Shiels AB, Knight T.. 2020. Local adaptation constrains drought tolerance in a tropical foundation tree. J Ecol. 108:1540–1552.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
