PLIP 2021: expanding the scope of the protein-ligand interaction profiler to DNA and RNA
- PMID: 33950214
- PMCID: PMC8262720
- DOI: 10.1093/nar/gkab294
PLIP 2021: expanding the scope of the protein-ligand interaction profiler to DNA and RNA
Abstract
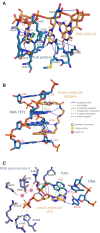
With the growth of protein structure data, the analysis of molecular interactions between ligands and their target molecules is gaining importance. PLIP, the protein-ligand interaction profiler, detects and visualises these interactions and provides data in formats suitable for further processing. PLIP has proven very successful in applications ranging from the characterisation of docking experiments to the assessment of novel ligand-protein complexes. Besides ligand-protein interactions, interactions with DNA and RNA play a vital role in many applications, such as drugs targeting DNA or RNA-binding proteins. To date, over 7% of all 3D structures in the Protein Data Bank include DNA or RNA. Therefore, we extended PLIP to encompass these important molecules. We demonstrate the power of this extension with examples of a cancer drug binding to a DNA target, and an RNA-protein complex central to a neurological disease. PLIP is available online at https://plip-tool.biotec.tu-dresden.de and as open source code. So far, the engine has served over a million queries and the source code has been downloaded several thousand times.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Soliman C., Yuriev E., Ramsland P.A.. Antibody recognition of aberrant glycosylation on the surface of cancer cells. Curr. Opin. Struct. Biol. 2017; 44:1–8. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

