Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike mutation T478K
- PMID: 33951211
- PMCID: PMC8242375
- DOI: 10.1002/jmv.27062
Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike mutation T478K
Abstract
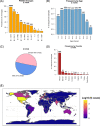
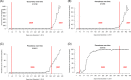
Several severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants have emerged, posing a renewed threat to coronavirus disease 2019 containment and to vaccine and drug efficacy. In this study, we analyzed more than 1,000,000 SARS-CoV-2 genomic sequences deposited up to April 27, 2021, on the GISAID public repository, and identified a novel T478K mutation located on the SARS-CoV-2 Spike protein. The mutation is structurally located in the region of interaction with human receptor ACE2 and was detected in 11,435 distinct cases. We show that T478K has appeared and risen in frequency since January 2021, predominantly in Mexico and the United States, but we could also detect it in several European countries.
Keywords: COVID-19; S:T478K; SARS-CoV-2; Spike; Spike:T478K; T478K; genomic surveillance.
© 2021 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures

Similar articles
-
Haplotype distribution of SARS-CoV-2 variants in low and high vaccination rate countries during ongoing global COVID-19 pandemic in early 2021.Infect Genet Evol. 2022 Jan;97:105164. doi: 10.1016/j.meegid.2021.105164. Epub 2021 Nov 27. Infect Genet Evol. 2022. PMID: 34848355 Free PMC article.
-
Emergence and spread of the potential variant of interest (VOI) B.1.1.519 of SARS-CoV-2 predominantly present in Mexico.Arch Virol. 2021 Nov;166(11):3173-3177. doi: 10.1007/s00705-021-05208-6. Epub 2021 Aug 27. Arch Virol. 2021. PMID: 34448936 Free PMC article.
-
V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity.J Virol. 2021 Jul 26;95(16):e0061721. doi: 10.1128/JVI.00617-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34105996 Free PMC article.
-
SARS-CoV-2 Infection: New Molecular, Phylogenetic, and Pathogenetic Insights. Efficacy of Current Vaccines and the Potential Risk of Variants.Viruses. 2021 Aug 25;13(9):1687. doi: 10.3390/v13091687. Viruses. 2021. PMID: 34578269 Free PMC article. Review.
-
Current understanding of the adaptive evolution of the SARS-CoV-2 genome.Yi Chuan. 2025 Feb;47(2):211-227. doi: 10.16288/j.yczz.24-231. Yi Chuan. 2025. PMID: 39924701 Review.
Cited by
-
Host Response to SARS-CoV2 and Emerging Variants in Pre-Existing Liver and Gastrointestinal Diseases.Front Cell Infect Microbiol. 2021 Oct 25;11:753249. doi: 10.3389/fcimb.2021.753249. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34760721 Free PMC article. Review.
-
Clinical features of children with coronavirus disease 2019 caused by Delta variant infection.Zhongguo Dang Dai Er Ke Za Zhi. 2021 Dec 15;23(12):1267-1270. doi: 10.7499/j.issn.1008-8830.2110043. Zhongguo Dang Dai Er Ke Za Zhi. 2021. PMID: 34911611 Free PMC article. Chinese, English.
-
Evolution of SARS-CoV-2 Variants: Implications on Immune Escape, Vaccination, Therapeutic and Diagnostic Strategies.Viruses. 2023 Apr 10;15(4):944. doi: 10.3390/v15040944. Viruses. 2023. PMID: 37112923 Free PMC article. Review.
-
SARS-CoV-2 Omicron Variant: Epidemiological Features, Biological Characteristics, and Clinical Significance.Front Immunol. 2022 Apr 29;13:877101. doi: 10.3389/fimmu.2022.877101. eCollection 2022. Front Immunol. 2022. PMID: 35572518 Free PMC article. Review.
-
Differences in Clinical Characteristics Between Delta Variant and Wild-Type SARS-CoV-2 Infected Patients.Front Med (Lausanne). 2022 Jan 3;8:792135. doi: 10.3389/fmed.2021.792135. eCollection 2021. Front Med (Lausanne). 2022. PMID: 35047533 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

