NMR illuminates intrinsic disorder
- PMID: 33951592
- PMCID: PMC8530845
- DOI: 10.1016/j.sbi.2021.03.015
NMR illuminates intrinsic disorder
Abstract
Nuclear magnetic resonance (NMR) has long been instrumental in the characterization of intrinsically disordered proteins (IDPs) and intrinsically disordered regions (IDRs). This method continues to offer rich insights into the nature of IDPs in solution, especially in combination with other biophysical methods such as small-angle scattering, single-molecule fluorescence, electron paramagnetic resonance (EPR), and mass spectrometry. Substantial advances have been made in recent years in studies of proteins containing both ordered and disordered domains and in the characterization of problematic sequences containing repeated tracts of a single or a few amino acids. These sequences are relevant to disease states such as Alzheimer's, Parkinson's, and Huntington's diseases, where disordered proteins misfold into harmful amyloid. Innovative applications of NMR are providing novel insights into mechanisms of protein aggregation and the complexity of IDP interactions with their targets. As a basis for understanding the solution structural ensembles, dynamic behavior, and functional mechanisms of IDPs and IDRs, NMR continues to prove invaluable.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of interest statement Nothing declared.
Figures
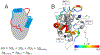
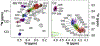
References
-
- Kakeshpour T, Ramanujam V, Barnes CA, Shen Y, Ying J, Bax A: A lowly populated, transient β-sheet structure in monomeric Aβ1-42 identified by multinuclear NMR of chemical denaturation. Biophys Chem 2021, 270:106531. - PMC - PubMed
-
Application of chemical denaturant titrations and chemical shift-based structure determination to identify a transient, weakly populated β-sheet in the Alzheimer’s Aβ1-42 peptide.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

