Methylated markers accurately distinguish primary central nervous system lymphomas (PCNSL) from other CNS tumors
- PMID: 33952317
- PMCID: PMC8097855
- DOI: 10.1186/s13148-021-01091-9
Methylated markers accurately distinguish primary central nervous system lymphomas (PCNSL) from other CNS tumors
Abstract
Background: Definitive diagnosis of primary central nervous system lymphoma (PCNSL) requires invasive surgical brain biopsy, causing treatment delays. In this paper, we identified and validated tumor-specific markers that can distinguish PCNSL from other CNS tumors in tissues. In a pilot study, we tested these newly identified markers in plasma.
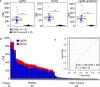
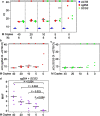
Results: The Methylation Outlier Detector program was used to identify markers in TCGA dataset of 48 diffuse large B-cell lymphoma (DLBCL) and 656 glioblastomas and lower-grade gliomas. Eight methylated markers clearly distinguished DLBCL from gliomas. Marker performance was verified (ROC-AUC of ≥ 0.989) in samples from several GEO datasets (95 PCNSL; 2112 other primary CNS tumors of 11 types). Next, we developed a novel, efficient assay called Tailed Amplicon Multiplexed-Methylation-Specific PCR (TAM-MSP), which uses two of the methylation markers, cg0504 and SCG3 triplexed with ACTB. FFPE tissue sections (25 cases each) of PCNSL and eight types of other primary CNS tumors were analyzed using TAM-MSP. TAM-MSP distinguished PCNSL from the other primary CNS tumors with 100% accuracy (AUC = 1.00, 95% CI 0.95-1.00, P < 0.001). The TAM-MSP assay also detected as few as 5 copies of fully methylated plasma DNA spiked into 0.5 ml of healthy plasma. In a pilot study of plasma from 15 PCNSL, 5 other CNS tumors and 6 healthy individuals, methylation in cg0504 and SCG3 was detectable in 3/15 PCNSL samples (20%).
Conclusion: The Methylation Outlier Detector program identified methylated markers that distinguish PCNSL from other CNS tumors with accuracy. The high level of accuracy achieved by these markers was validated in tissues by a novel method, TAM-MSP. These studies lay a strong foundation for a liquid biopsy-based test to detect PCNSL-specific circulating tumor DNA.
Keywords: Circulating tumor DNA; DNA methylation; Epigenetic biomarkers; Liquid biopsy; Primary central nervous system lymphoma; TAM-MSP.
Conflict of interest statement
S.S. has received research grants, licensing/royalty fees and consulting fees from Cepheid for the QM-MSP assays. No other authors have disclosed any conflicts of interest.
Figures



References
-
- Batchelor TT, DeAngelis LM. Lymphoma and leukemia of the nervous system. New York: Springer; 2013.
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. - PubMed
-
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–1947. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

