FoldAffinity: binding affinities from nDSF experiments
- PMID: 33953265
- PMCID: PMC8099913
- DOI: 10.1038/s41598-021-88985-z
FoldAffinity: binding affinities from nDSF experiments
Abstract
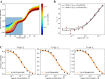
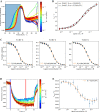
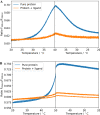
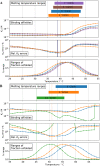
Differential scanning fluorimetry (DSF) using the inherent fluorescence of proteins (nDSF) is a popular technique to evaluate thermal protein stability in different conditions (e.g. buffer, pH). In many cases, ligand binding increases thermal stability of a protein and often this can be detected as a clear shift in nDSF experiments. Here, we evaluate binding affinity quantification based on thermal shifts. We present four protein systems with different binding affinity ligands, ranging from nM to high μM. Our study suggests that binding affinities determined by isothermal analysis are in better agreement with those from established biophysical techniques (ITC and MST) compared to apparent Kds obtained from melting temperatures. In addition, we describe a method to optionally fit the heat capacity change upon unfolding ([Formula: see text]) during the isothermal analysis. This publication includes the release of a web server for easy and accessible application of isothermal analysis to nDSF data.
Conflict of interest statement
The authors declare no competing interests.
Figures
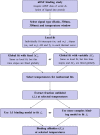
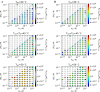
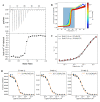
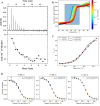
References
Publication types
LinkOut - more resources
Full Text Sources

