The human memory T cell compartment changes across tissues of the female reproductive tract
- PMID: 33953338
- PMCID: PMC8225572
- DOI: 10.1038/s41385-021-00406-6
The human memory T cell compartment changes across tissues of the female reproductive tract
Abstract
Memory CD4 T cells in tissues fulfill numerous functions that are critical for local immune homeostasis and protection against pathogens. Previous studies have highlighted the phenotypic and functional heterogeneity of circulating and tissue-resident memory CD4 T cells across different human tissues such as skin, lung, liver, and colon. Comparatively little is known in regard to memory CD4 T cells across tissues of the female reproductive tract (FRT). We examined CD4 T cells in donor-matched vaginal, ecto- and endocervical tissues, which differ in mucosal structure and exposure to external environmental stimuli. We hypothesized that this could be reflected by tissue-specific differences in the memory CD4 T cell compartment. We found differences in CD4 subset distribution across these tissues. Specifically, CD69+CD103+ CD4 T cells were significantly more abundant in vaginal than cervical tissues. In contrast, the transcriptional profiles of CD4 subsets were fairly conserved across FRT tissues. CD69+CD103+ CD4 T cells showed a TH17 bias independent of tissue niche. Our data suggest that FRT tissues affect T cell subset distribution but have limited effects on the transcriptome of each subset. We discuss the implications for barrier immunity in the FRT.
Conflict of interest statement
Conflict of Interest Statement
The authors declare no conflict.
Figures
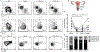

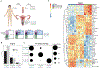




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

