Delayed production of neutralizing antibodies correlates with fatal COVID-19
- PMID: 33953384
- PMCID: PMC8785364
- DOI: 10.1038/s41591-021-01355-0
Delayed production of neutralizing antibodies correlates with fatal COVID-19
Erratum in
-
Author Correction: Delayed production of neutralizing antibodies correlates with fatal COVID-19.Nat Med. 2021 Jul;27(7):1309. doi: 10.1038/s41591-021-01416-4. Nat Med. 2021. PMID: 34145437 Free PMC article. No abstract available.
Abstract
Recent studies have provided insights into innate and adaptive immune dynamics in coronavirus disease 2019 (COVID-19). However, the exact features of antibody responses that govern COVID-19 disease outcomes remain unclear. In this study, we analyzed humoral immune responses in 229 patients with asymptomatic, mild, moderate and severe COVID-19 over time to probe the nature of antibody responses in disease severity and mortality. We observed a correlation between anti-spike (S) immunoglobulin G (IgG) levels, length of hospitalization and clinical parameters associated with worse clinical progression. Although high anti-S IgG levels correlated with worse disease severity, such correlation was time dependent. Deceased patients did not have higher overall humoral response than discharged patients. However, they mounted a robust, yet delayed, response, measured by anti-S, anti-receptor-binding domain IgG and neutralizing antibody (NAb) levels compared to survivors. Delayed seroconversion kinetics correlated with impaired viral control in deceased patients. Finally, although sera from 85% of patients displayed some neutralization capacity during their disease course, NAb generation before 14 d of disease onset emerged as a key factor for recovery. These data indicate that COVID-19 mortality does not correlate with the cross-sectional antiviral antibody levels per se but, rather, with the delayed kinetics of NAb production.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
A.I. served as a consultant for Spring Discovery, Boehringer Ingelheim and Adaptive Biotechnologies. I.Y. reports being a member of the mRNA-1273 Study Group and has received funding to her institution to conduct clinical research from BioFire, MedImmune, Regeneron, PaxVax, Pfizer, GlaxoSmithKline, Merck, Novavax, Sanofi-Pasteur and Micron. All other authors declare no competing financial interests.
Figures

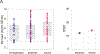

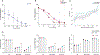





References
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007517/AI/NIAID NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- T03 OH008607/OH/NIOSH CDC HHS/United States
- K24 AG042489/AG/NIA NIH HHS/United States
- NIAID 2T32AI007517/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Medical

