ZebraShare: a new venue for rapid dissemination of zebrafish mutant data
- PMID: 33954026
- PMCID: PMC8051354
- DOI: 10.7717/peerj.11007
ZebraShare: a new venue for rapid dissemination of zebrafish mutant data
Abstract
Background: In the past decade, the zebrafish community has widely embraced targeted mutagenesis technologies, resulting in an abundance of mutant lines. While many lines have proven to be useful for investigating gene function, many have also shown no apparent phenotype, or phenotypes not of interest to the originating lab. In order for labs to document and share information about these lines, we have created ZebraShare as a new resource offered within ZFIN.
Methods: ZebraShare involves a form-based submission process generated by ZFIN. The ZebraShare interface (https://zfin.org/action/zebrashare) can be accessed on ZFIN under "Submit Data". Users download the Submission Workbook and complete the required fields, then submit the completed workbook with associated images and captions, generating a new ZFIN publication record. ZFIN curators add the submitted phenotype and mutant information to the ZFIN database, provide mapping information about mutations, and cross reference this information across the appropriate ZFIN databases. We present here examples of ZebraShare submissions, including phf21aa, kdm1a, ctnnd1, snu13a, and snu13b mutant lines.
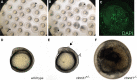
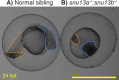
Results: Users can find ZebraShare submissions by searching ZFIN for specific alleles or line designations, just as for alleles submitted through the normal process. We present several potential examples of submission types to ZebraShare including a phenotypic mutants, mildly phenotypic, and early lethal mutants. Mutants for kdm1a show no apparent skeletal phenotype, and phf21aa mutants show only a mild skeletal phenotype, yet these genes have specific human disease relevance and therefore may be useful for further studies. The p120-catenin encoding gene, ctnnd1, was knocked out to investigate a potential role in brain development or function. The homozygous ctnnd1 mutant disintegrates during early somitogenesis and the heterozygote has localized defects, revealing vital roles in early development. Two snu13 genes were knocked out to investigate a role in muscle formation. The snu13a;snu13b double mutant has an early embryonic lethal phenotype, potentially related to a proposed role in the core splicing complex. In each example, the mutants submitted to ZebraShare display phenotypes that are not ideally suited to their originating lab's project directions but may be of great relevance to other researchers.
Conclusion: ZebraShare provides an opportunity for researchers to directly share information about mutant lines within ZFIN, which is widely used by the community as a central database of information about zebrafish lines. Submissions of alleles with a phenotypic or unexpected phenotypes is encouraged to promote collaborations, disseminate lines, reduce redundancy of effort and to promote efficient use of time and resources. We anticipate that as submissions to ZebraShare increase, they will help build an ultimately more complete picture of zebrafish genetics and development.
Keywords: Collaboration; Zebrafish; ctnnd1; kdm1a; lsd1; nhp2l1; phf21a; snu13.
©2021 DeLaurier et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures




References
-
- Alharatani R, Ververi A, Beleza Meireles A, Ji W, Mis E, Patterson QT, Griffin JN, Bhujel N, Chang CA, Dixit A, Konstantino M, Healy C, Hannan S, Neo N, Cash A, Li D, Bhoj E, Zackai EH, Cleaver R, Baralle D, McEntagart M, Newbury-Ecob R, Scott R, Hurst JA, Au PYB, Hosey MT, Khokha M, Marciano DK, Lakhani SA, Liu KJ. Novel truncating mutations in CTNND1 cause a dominant craniofacial and cardiac syndrome. Human Molecular Genetics. 2020;29:1900–1921. doi: 10.1093/hmg/ddaa050. - DOI - PMC - PubMed
-
- Anderson JL, Mulligan TS, Shen M-C, Wang H, Scahill CM, Tan FJ, Du SJ, Busch-Nentwich EM, Farber SA. mRNA processing in mutant zebrafish lines generated by chemical and CRISPR-mediated mutagenesis produces unexpected transcripts that escape nonsense-mediated decay. PLOS Genetics. 2017;13:e1007105. doi: 10.1371/journal.pgen.1007105. - DOI - PMC - PubMed
-
- Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SAT, Paddock H, Pich C, Ramachandran S, Ruef BJ, Ruzicka L, Schaper HBauer, Schaper K, Shao X, Singer A, Sprague J, Sprunger B, Van Slyke C, Westerfield M. ZFIN: enhancements and updates to the zebrafish model organism database. Nucleic Acids Research. 2011;39:D822–D829. doi: 10.1093/nar/gkq1077. - DOI - PMC - PubMed
-
- Chong JX, Yu J-H, Lorentzen P, Park KM, Jamal SM, Tabor HK, Rauch A, Saenz MS, Boltshauser E, Patterson KE, Nickerson DA, Bamshad MJ. Gene discovery for Mendelian conditions via social networking: de novo variants in KDM1A cause developmental delay and distinctive facial features. Genetics in Medicine. 2016;18:788–795. doi: 10.1038/gim.2015.161. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

